![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
60 Cards in this Set
- Front
- Back
|
replication
|
DNA synthesis
|
|
|
transcription
|
RNA synthesis
|
|
|
translation
|
protein synthesis
|
|
|
In prokaryotes (organisms without nuclei such as bacteria) replication occurs in the _____.
|
cytoplasm
|
|
|
In eukaryotes (organisms with nuclei such as humans) replication occurs in the _____ during S phase of the cell cycle.
|
nucleus
|
|
|
The purpose for DNA replication is to duplicate _____, so that after mitosis each daughter cell will inherit a complete genome.
|
chromosomes
|
|
|
DNA replication only occurs in dividing cells. Non-dividing cells blocked in ___ do not progress to ___ phase, so they do not replicate their DNA.
|
Go, S
|
|
|
Characteristics of DNA Replication: Requirements
|
DNA Polymerase, Mg+2, Template, Primer, dNTPs
|
|
|
Characteristics of DNA Replication: Direction
|
5'-3'
|
|
|
Characteristics of DNA Replication: Complimentary
|
For each A of the template strand, a T is added to the new strand, and each G is matched with a C.
|
|
|
Characteristics of DNA Replication: Proofreading
|
3' to 5' exonuclease activity removes mismatched nucleotides.
|
|
|
Characteristics of DNA Replication: Bidirectional
|
Replication proceeds in both directions from central origins of replication (ori).
|
|
|
Characteristics of DNA Replication: Discontinuous
|
The lagging strand is synthesized as short stretches.
|
|
|
Characteristics of DNA Replication: Semiconserative
|
One strand of a daughter chromosome is newly synthesized, the other template strand is inherited intact from the parental chromosome.
|
|
|
DNA Polymerase (Pol#)
|
An enzyme that catalyzes polymerization of dNTPs into DNA.
|
|
|
What are the prokaryotic DNA polymerases?
|
Pol I, II & III (I, II & III are also RNA Pols)
|
|
|
What are the eukaryotic DNA polymerases?
|
Pol δ, α, β & γ
|
|
|
DNA Pol δ
|
-Location: nucleus
-Fnx: leading strand synth -Processivity: >100,000 bp -Proofreading: + -RNA primer: + |
|
|
DNA Pol α
|
-Location: nucleus
-Fnx: lagging strand synth -Processivity: ~180 bp -Proofreading: - -RNA primer: + |
|
|
DNA Pol β
|
-Location: nucleus
-Fnx: repair gap filling -Processivity: ~20 bp -Proofreading: - -RNA primer: - |
|
|
DNA Pol γ
|
-Location: mitochondria
-Fnx: synth of both strands -Processivity: >8300 bp -Proofreading: + -RNA primer: - |
|
|
Although the ___ ion is not incorporated into DNA, it is a cofactor required for DNA polymerase activity.
|
Mg2+
|
|
|
To synthesize a new strand, DNA polymerase must read a preexisting strand. This preexisting strand is known as the _____.
|
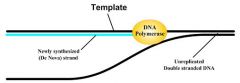
Template
|
|
|
Deoxyribonucleotides (dNTPs)
|
building blocks of DNA. All four are required for replication.
|
|
|
Remember that dNTPs lack _____ (hence the prefix deoxy). Thymine, which has a methyl group, is used instead of methyl free _____, which is used for RNA.
|
2' hydroxyls, uridine
|
|
|
Hydrolysis of the _____ provides the energy required for nucleotide polymerization.
|
triphosphate bonds; Energy is stored in the triphosphate bonds as electrostatic repulsion of their negatively charged oxygens.
|
|
|
The final requirement for DNA replication is a primer. This a free hydroxyl (OH) at the ___ position of the terminal nucleotide of a previously existing strand. A strand with a free ___ OH is also referred to as a primer.
|
3', 3'; The reason for this requirement is DNA polymerase can only bind a 3' OH with the 5’ phosphate of an incoming nucleotide. The opposite reaction will not occur. The hydroxyl of a new nucleotide cannot be bound with the phosphate of a preexisting strand
|
|
|
DNA is synthesized in the 5'-3' direction, therefore the template strand must be read _____.
|
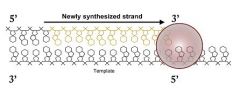
3'-5'
|
|
|
Pol __ and Pol ___ have the abilit.y to “double check” whether each new nucleotide is complimentary to the corresponding base of the template.
|
δ, γ; If an incorrect nucleotide is incorporated, the polymerase’s 3'-5' exonuclease activity “kicks back,” excising the mismatch. Pol α and Pol β lack this activity.
|
|
|
How is possible for the lagging strand to be synthesized in the 3'-5' direction if no DNA polymerase is capable of doing so?
|
Okazaki fragments; The dilemma is resolved by discontinuously synthesizing the lagging strand as short stretches (100-200 bp), in the correct 5 '- 3' direction. These stretches are known as Okazaki fragments.
|
|
|
Okazaki fragments
|
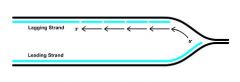
|
|
|
Semiconservative- One strand of a daughter chromosome is newly synthesized, the other template strand is inherited intact from the parental chromosome.
|
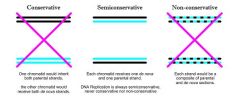
|
|
|
origin of replication
(ori) |
Sequences where DNA replication begins. Factors recognize and bind the specific ori sequence, unwind its DNA, and attract the components of the replication apparatus, which moves on down the chromosome replicating DNA as it goes.
|
|
|
How many ORIs do prokaryotes and eukaryotes have?
|
Proks 1, Euks multiple; this is how eukaryotic DNA replication takes only a little more time than prokaryotic, even though our genome is 1000s of times larger
|
|
|
How can eukaryotic DNA replication be bidirectional?
|
two replication apparatuses actually assemble around the ori and move in opposite directions
|
|
|
replicon- region of a eukaryotic chromosome that is replicated as a unit, from one central ori.
|
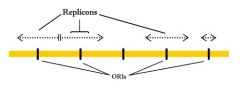
Replication begins at the ori in the center of a replicon, and extends in both directions until it reaches the end of an adjacent replicon. Because there are multiple replicons, different regions of the genome are replicated simultaneously.
|
|
|
What is the average length of our replicons? (BPs)
|
about 200 kb. This is the approximate length of DNA loops anchored to scaffold proteins, suggesting that these proteins may be involved in defining replicon length.
|
|
|
helicase
|
-factor which unwinds DNA
-binds to an ori where it opens the double helix so DNA polymerases can replicate the strands. -part of replication apparatus |
|
|
single stranded binding proteins (SSB)
|
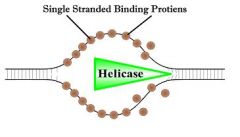
Factors that stabilize single stranded DNA by preventing it from winding back into a double helix.
|
|
|
DNA replication apparatus
|
-cluster of factors assembles around helicase to replicate DNA
-Two apparatuses assemble, one on each side of the ori, and each moves in opposite directions, replicating DNA as they go |
|
|
What factors are part of the DNA replication apparatus?
|
-helicase
-Pol δ -Pol α -β clamp -primase |
|
|
Are SSBs part of the DNA replication apparatus?
|
nope
|
|
|
β clamp
|
ring like protein that wraps around DNA to stabilize the association of the replication apparatus
|
|
|
β clamp is required for Pol __ processivity.
|
pol δ; Without the clamp, Pol δ only replicates short oligonucleotides (< 200 bp). With the clamp, stretches greater than 100 kb are produced.
|
|
|
Pol __ is the enzyme which replicates the leading strand in the ____ direction.
|
Pol δ, 5'-3'; It reads the template one base at a time, incorporating complimentary nucleotides, and ligating their 5' phosphate to the 3' OH of the growing leading strand.
|
|
|
As helicase unwinds DNA, single strand binding proteins keep the lagging strand template unwound.
|
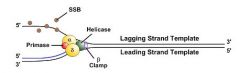
|
|
|
primase
|
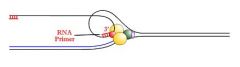
enzyme binds the unwound lagging strand template and transcribes a short stretch of RNA (<15 bp). This RNA serves as a primer, providing the 3' OH group required by Pol α.
|
|
|
DNA polymerase α (Pol α)
|
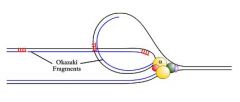
enzyme uses the RNA primer to synthesize an Okazaki fragment of the lagging strand.
|
|
|
Does Pol α have proofreading capability?
|
no, suprisingly
|
|
|
Why is the lagging strand held in a loop?
|
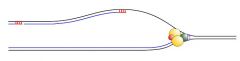
as Pol α synthesizes one Okazaki fragment, helicase continues unwinding single stranded DNA for the next fragment. Therefore, the lagging strand is held in a loop.
|
|
|
What happens to the okazaki fragments?
|
Pol α continues replicating the Okazaki fragment until it reaches the primer at the end of the previous fragment. The lagging strand is then released, primase makes the next primer at the end of the new single stranded region, and the process is repeated.
|
|
|
Removal of RNA Primers from the Lagging Strand
|
After the replication apparatus passes, it leaves a lagging strand full of RNA primers which must be removed. This involves three separate enzymes:
-RNAase -DNA Pol β -DNA ligase |
|
|
RNAase
|
This enzyme digests any RNA.
In replication, it serves to remove the lagging strand primers. |
|
|
DNA polymerase β (Pol β)
|
fills in gaps in DNA, about 20 bp long.
In replication, it fills in the gaps left after the RNA primers are removed. Note, DNA polymerases can only add nucleotides; they cannot link DNA fragments. Therefore, Pol β leaves nicks in the DNA. |
|
|
DNA ligase
|
enzyme binds any free 3’ hydroxyls and 5 phosphates of DNA.
In replication, it seals the nicks between the Okazaki fragments left by Pol β. |
|
|
topoisomerase
|
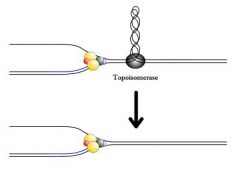
A consequence of unwinding by helicase is that supercoiling increases. Topoisomerases serve to restore the DNA to the proper level of supercoiling.
|
|
|
What happens to the overhang left on the end of the chromosome when the last RNA primer is removed?
|
Telomerase fills in the gap; it's a reverse transcriptase which extends the length of telomeres.
|
|
|
What would happen without telomerase?
|
If this dilemma were not resolved, then every time dividing cells replicated their DNA, the lagging strands would loss a little bit of telomeric sequence, until their telomeres were completely whittled away. This would destabilize the chromosome.
|
|
|
What does telomerase have to do with aging?
|
-Telomerase activity decreases with age, as does the length of our telomeres.
-Primary cell culture lines divide a limited number of times & then die. This is believed to be because they have no telomerase activity. -In immortalized cell culture lines telomerase is activated, so they can divide indefinitely. |
|
|
What does telomerase have to do with cancer?
|
Telomerase is usually activated in cancer cells, and this is believed to contribute to their immortalization
|

