![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
38 Cards in this Set
- Front
- Back
|
polymerization of nucleotides
|
to make a dinucleotide, the phosphate is esterified to the 3'OH of another nucleotide
= phosphodiester linkage directionality: 5' --> 3' (left to right) can write as: 5'-ATG-3' or: p-ATG-OH |
|
|
how does DNA half-life compare to RNA?
|
RNA t1/2 = days
DNA t1/2 - thousands of years -difference is OH in 2' position (self-destruct mechanism) -OH is close to phosphorous atom and can cleave off (unstable) =Alkaline Hydrolysis of RNA |
|
|
DNA structure
|
-double helix
-2 polynucleotides associated by H-bonding between the bases -2 polynucleotide ("DNA strands") are complementary -the sequence of one determines the sequence of the other (and vice versa) -the 2 strands are anti-parallel (they run in opposite directions) |
|
|
why does G-C and A-T?
|
-if mix G&C together, exothermic rxn (spontaneous)
-in G-C complex, 3 complementary H-bonds that can form: donors & acceptors = molecular recognition (perfect fit in terms of distances and angles) -the same for A-T (2 H-bonds) |
|
|
Forces that determine DNA structure
|
1) Electrostatic (repulsion)
-work against Watson-Crik structure 2) Hydrophobic "forces" -limit exposure or nitrogenous bases from aqueous conditions -work for structure 3) Hydrogen bonds -work for structure 4) Stacking interactions -aromatic structures (nitrogenous bases) stack on top of each other -work for structure |
|
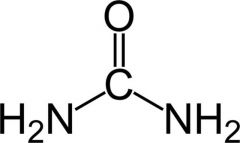
How might urea affect DNA stability?
|
-lots of H-bond capabilities
-can compete w H-bonds and interfere w DNA -destabilize DNA |
|
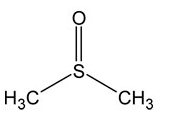
How might DNSO affect DNA stability?
|
-mostly non-polar (methyl groups) w polar part
-messes up hydrophobic forces -interfere w hydrophobic effect of DNA -destabilizes DNA |
|
why is binding between complementary PNA/DNA stronger than DNA/DNA?
|
-backbone has no negative charge
-do not have strand-strand repulsion w DNA (stabilizes double strand PNA-DNA) -in DNA-DNA, both are negative so get repulsion |
|
|
what is the effect of changing ionic strength on DNA stability?
|
-as increase [salt] it shields neg. charge on phosphate, increasing stability
-too high [salt] -> get salting out (no more water which is needed for hydrophobic effect of DNA) -> decreasing stability |
|
|
B form of DNA
|
10.5 residues per turn of helix
1 turn is 36A in length ~20A diameter 3.5A helix rise per base pair 6 degree base tilt normal to helical axis C-2' endo sugar pucker anti glycosyl bond conformation right handed has major groove and minor groove |
|
|
what is the conformation of base w.r.t. pentose ring in purines
|
can do syn or anti, but anti more stable
|
|
|
what is the conformation of base w.r.t. pentose ring in pyrimidines
|
can only do anti
|
|
|
what are the differences between the major and minor grooves?
|
major groove is larger and contains more infor
-H-bond donors/acceptors, methyl groups (Thymine) -major groove s site of most protein-DNA interactions |
|
|
what is the pattern for G-C pair in major groove?
|
Acceptor, Acceptor, Donor
|
|
|
what is the pattern for G-C pair in minor groove?
|
Acceptor, Donor, Acceptor
|
|
|
what is the pattern for A-T pair in major groove?
|
Acceptor, Donor, Acceptor, Methyl
|
|
|
what is the pattern for A-T pair in minor groove?
|
Acceptor, Donor
|
|
|
DNA denaturation
|
-can be denatured by temp and pH
-disruption of H-bond and hydrophobic interactions between the bases -results in separation of strands ds (native) <--> ss (denatured) -viscosity changes (ds more viscous) -UV abs changes |
|
|
what is melting temp (Tm)
|
the temperature at the midpoint in the denaturation curve
|
|
|
influences on Tm
|
1) increase G-C content, increase Tm
2) increase [salt] decrease Tm (salting out) 3) low (<2.3) or high (>11.5) pH, decrease Tm 4) organic compounds: -increase [urea], decrease Tm -increase [lysine], increase Tm -increase [glutamate], increase Tm (lys (-) and glu (+) have charge that can shield neg. charge on phosphate) |
|
|
DNA renaturation steps
|
1) nucleation
-the 2 strands find a region of complementarity and form a short double helix 2) zippering -in either direction from the paired region of complementarity, the double helix is elongated |
|
|
annealing temp
|
temp where strands can find each other and get nucleation
= Tm - 25 deg C |
|
|
supercoiling
|
DNA is being used to make RNA
-process of separating strands ahead = stress is overwound behind = stress is underwound onderwound: > 10.5 bases/turn (looser) overwound: < 10.5 bases/turn (tighter) relaxed = 10.5 being over/underwound causes supercoiling b/c it wants to be 10.5 which is the most stable form |
|
|
supercoiling in prokaryotes
|
keep DNA in underwound state
-easier to do strand separation needed for replication -makes DNA more compact |
|
|
2 forms of supercoil:
|
1) plectomeme
-twisted sheet 2) solemoid -1 loop |
|
|
linking number
|
the number of crosses a single strand makes across the other
-2 rings linked together, Lk = 1 -if ring is not closed, Lk = undefined in relaxed state: Lk0 = bp/10.5 |
|
|
Agarose Gel Electrophoresis
|
-utilizes polysaccharide matrix w large pores
-neg. charge (from phosphates) is proportional to length of DNA -separation based on size -if relaxed and supercoiled DNA of same size, would get 2 bands (relaxed will travel shorter distance, smaller will travel farther) -supercoiling reduces the effective size of the molecule, allowing it to migrate faster -each band represents a discrete linking number |
|
|
Topoisomerases
|
move DNA to relaxed state
to change Lk: 1) have to make a cut 2) change 3) seal 2 types: topo I and topo II |
|
|
Topoisomerase I mechanism
|
change Lk by 1
does not req ATP 1) active site Tyr attacks a phosphodiester bond in 1 DNA strand, cleaving it & creating a covalent 5'-phosphotyrosyl protein-DNA linkage 2) enzyme changes to open conformation 3) unbroken DNA strand passes through break in first strand 4) enzyme in closed conf; liberated 3'OH attacks the 5'-phosphotyrosyl protein-DNA linkage to religate the cleaved DNA strand |
|
|
Topoisomerase II mechanism
|
changes Lk by 2
requires ATP 1) multisubunit enzyme binds a segment of a DNA molecule 2) a 2nd segment of same DNA molecule is bound at the N gate 3) 2nd DNA is trapped. 1st piece cleaved on both strands to form 2 5'-phosphotyrosyl linkages to the enzyme 4) 2nd DNA segment passed through break 5) broken DNA religated & 2nd piece released through the C gate |
|
|
bacterial gyrase
|
special type of topo II
fxn: underwinds bacterial DNA (need ATP to take DNA from relaxed to underwound) |
|
|
sulfolobus acidocaldarius grows optimally at 75-80C and pH = 2-3, under strictly aerobic conditions
-DNA typically denatures at those temps, what is the mechanism to protect against denaturation? |
-high concentration of G and C (75%)
-overwinds the DNA (special gyrase that uses ATP to overwind DNA and stabilizes it at high temp) |
|
|
how is bacterial supercoiling maintained?
|
bacterial supercoiling levels are determined by the balance between the activity of the topoisomerases
|
|
|
how is eukaryotic supercoiling maintained?
|
by binding to histones and relaxation of DNA between nucleosomes
|
|
|
chromosome structure in prokaryotes
|
E coli chromosome:
-4.5 mbp of DNA in a single circle -the chromosome is highly compacted -~500 supercoiled loops -histone-like proteins (HU proteins) -nucleoid |
|
|
chromosome structure in eukaryotes
|
-very compact structures made of chromatin (DNA-protein complex)
-the basic unit is a nucleosome -the nucleosome can be packaged together into higher order structures |
|
|
properties of histones
|
-form octomer (2 of each histone)
-contain high amounts of Lys & Arg (basic aas have positive charge on nitrogen to go w negative charge on phosphate) -histone is core of nucleosomes w DNA wrapped around |
|
|
Telomeres
|
-then ends of linear chromosomes
-consist of tandem repeats of a short (usually 6-8NT) DNA sequence of T,G in one strand -form "T loop" w 3' overhang -prevents degradation on ends of chromosome |

