![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
28 Cards in this Set
- Front
- Back
|
Nucleotides
What are they? 3 components |
Monomers of the nucleic acids DNA and RNA
1) Nitrogenous base 2) Pentose sugar 3) Phosphate group |
|
|
How do DNA and RNA differ?
|
DNA
• Has Thymine, but no uracil • Pentose = deoxyribose RNA • Has Uracil, but no Thymine • Pentose = ribose |
|
|
Pyrimidines vs. purines
|
Pyrimidines
• smaller 6-membered ring • Cytosine, Uracil, Thymine (CUT) Purine • larger 6-membered ring joined to a 5-membered ring • Adenine, Guanine (AG) |
|
|
How do nitrogenous bases pair?
By what type of bond and how many? |
C-G = 3 hydrogen bonds
DNA: • A-T = 2 hydrogen bonds RNA: • A-U = 1 hydrogen bond Hydrogen bonding |
|
|
Do nucleotides have a positive or negative charge?
Acidic or basic? |
Negative and acidic due to phosphate group
|
|
|
What type of bonds join nucleotides?
|
Phosphodiester bonds
|
|
|
What is the structure of a pentose? 1
What is it composed of and where are these components located? 6 What is the growing end and why? 3 By which process does it grow? 1 Draw it! |
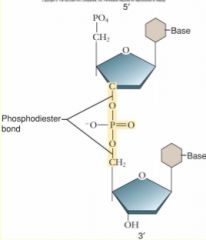
Structure
• 5-membered ring Composed of • 1' C is at right, count clockwise • 5' C branches off of 4' C • 5th ring member is an O • hydroxyl at 3' • phosphate at 5' • phosphodiester bonds between functional groups and C's Growing end at 3'!! • OH polarity = positive • PO4 polarity = negative • OH and PO4 are attracted to each other Process • Dehydration synthesis |
|
|
What about DNA was discovered through x-ray crystallography?
2 |
1) X-ray beams were diffracted when they hit DNA molecules to show that DNA is a double helix
2) Base pairs discerned from % of each base in the genome seen from the x-ray |
|
|
The orientation of DNA's backbone and bases
|
• Sugar-phosphate backbone is located on the outside towards the aqueous environment
• Nitrogenous bases located on the inside joining the 2 helixes by hydrogen bonds |
|
|
Antiparallel
|
DNA strands are complementary to one another = they run parallel but reverse direction
5' - 3' 3' - 5' Rather then: 5' - 3' 5' - 3' |
|
|
What are the 3 proposed models of DNA replication?
Which is correct? |
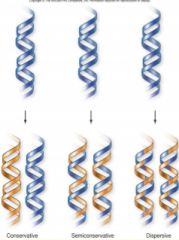
1) Conservative model = parental strands remain intact (conserved), and daughter strands are all new
2) Semiconservative model = parental strand is a template for replication; daughter contains 1 of the parental strands and 1 made from it 3) Dispersive model = daughter strands contains DNA of both parental and new daughter strand segments Semiconservative is correct |
|
|
Mutations
What is involved in correcting them? |
Errors in base pairing during DNA replication/synthesis
Enzymes |
|
|
Bacterial DNA replication
2 sites of importance |
1) Ori = origin of replication
2) Termination sequences These are sequences of DNA, not genes |
|
|
Replication bubbles
What would happen if there was just 1 replication bubble? |
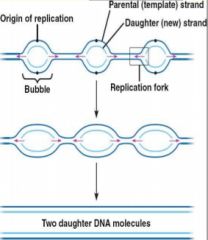
• There are ori's running down the length of each DNA strand
• replication begins at each ori, forming bubbles at each site • replication proceeds in opposite directions on each side of the bubble all adjacent bubbles meet and form 2 new strands • If Eukaryote DNA had only 1 replication site the process would take too long; multiple bubble speed things up! |
|
|
To polymerize...
|
is to grow through dehydration synthesis
Monomers → polymers |
|
|
DNA Polymerase III
What it does By which process What type of bond |
Polymerizes DNA by adding nucleotide monomers to the 3' end of a growing strand by dehydration synthesis
The strands are joined by phosphodiester bonds |
|
|
Where do the nucleotides that pair to the parental strand come from?
What are these nucleotides really? |
They are free nucleotides from the nucleoplasm
Nucleosides |
|
|
Nucleoside triphosphate
|
The free DNA monomers that DNA poly III adds to the 3' end of the growing strand
Consists of: • sugar • nitrogenous base • 3 phosphates |
|
|
Pyrophosphate
|
2 phosphates that are removed from nucleoside triphosphate when nucleoside triphosphate bonds to the growing DNA strand
|
|
|
What processes drives the growth of DNA strands?
|
Exergonic removal of pyrophosphate from one nucleoside triphosphate drives endergonic bonding of another nucleoside triphosphate
|
|
|
2 daughter strands
Difference in their construction |
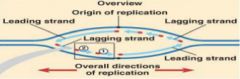
Half of each daughter is a leading strand and half is a lagging strand
Leading strand = adds nucleosides as it proceeds away from ori, into the replication fork Lagging strand = adds nucleosides as it proceeds towards ori, away from the fork Leading strand is replicated continuously Lagging strand is built in fragments |
|
|
Okazaki fragments
|
Short DNA fragments that build the lagging strand
|
|
|
Primers
How many are needed? Primase Why are they needed? |
Primers = Short RNA fragments which bond to the parental strand to act as a foundation for the daughter strand to be built upon
Primase = enzyme that joins primers to parental DNA strand Leading strand needs 1 placed at ori Lagging strand needs 1 per Okazaki fragment DNA poly III can only add new DNA nucleotides to preexisting nucleotides |
|
|
DNA Polymerase I
DNA ligase |
DNA Polymerase I = enzyme that replaces RNA primer with DNA nucleotides
DNA ligase = once the primers are replaced with DNA, this enzyme joins Okazaki fragments at their sugar-phosphate backbones |
|
|
What are the mechanics of the lagging strand?
4 |
1) An RNA primer is placed by primase behind the leading strand
2) An Okazaki fragment is laid down by DNA poly III in front of the primer (between primer and leading strand) 3) The primer is replaced with DNA nucleotides by DNA poly I 4) The Okazaki fragments are joined by DNA ligase |
|
|
Helicase
|
Unwinds the double helix
|
|
|
Single-strand binding proteins
|
Prevents single DNA strands from breaking when they have been unwound from the helix
|
|
|
Topoisomerase
|
DNA de-tangler
Cuts, unwinds, and reattaches DNA strands at each replication fork Think of teased hair - this prevents that |

