![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
259 Cards in this Set
- Front
- Back
|
1 Dalton=
|
1 AMU (atomic mass unit)= 1.7x10^-24 grams
|
|
|
element
|
Substance that cannot be broken down into other substances by chemical reactions.
|
|
|
compound
|
Substance consisting of two or more different elements combined in a fixed ratio.
|
|
|
How many elements are essential to life?
|
25
|
|
|
emergent properties
|
A compound has characteristics beyond those of its combined elements.
|
|
|
neutrons
|
Mass of 1 dalton.
Charge 0 (neutral) |
|
|
protons
|
Mass of 1 dalton.
Charge +1 |
|
|
electrons
|
Mass negligible.
Charge -1 |
|
|
Atomic number=
|
number of protons
|
|
|
Mass number=
|
protons + neutrons
|
|
|
isotopes
|
Have more neutrons than other atoms of the same element and therefore weigh more.
|
|
|
radioactive isotope
|
Nucleus decays spontaneously, giving off particles and energy.
|
|
|
valence electrons
|
Outer electrons. The chemical properties of an atom depend mostly on the number of valence electrons.
|
|
|
molecule
|
Two or more atoms held together by covalent bonds.
|
|
|
What is the difference between a molecule and a compound?
|
A molecule is formed when two or more atoms join together by covalent bonds. A compound is a molecule that combines at least two different elements. All compounds are molecules, but not all molecules are compounds.
|
|
|
Valence
|
Number of unpaired electrons in the valence shell; bonding capacity.
|
|
|
Valence of hydrogen
|
1
|
|
|
Valence of oxygen
|
2
|
|
|
Valence of nitrogen
|
3
|
|
|
Valence of carbon
|
4
|
|
|
Valence of phosphorus
|
5 in biologically important molecules. It forms three single bonds and one double bond.
|
|
|
electronegativity
|
Attraction of an atom for the electrons of a covalent bond. The more electronegative an atom, the more strongly it pulls shared electrons toward itself.
|
|
|
nonpolar covalent bond
|
Electrons are shared equally
|
|
|
polar covalent bond
|
One atom is more electronegative than the other. The electrons spend more time around the electronegative atom.
|
|
|
ionic bond
|
The more electronegative ion strips an electron completely away from its partner.
|
|
|
cation
|
A positively-charged ion
|
|
|
anion
|
A negatively-charged ion
|
|
|
hydrogen bond
|
A hydrogen atom covalently bonded to one electronegative atom is also attracted to another electronegative atom. Effective glue in large numbers. Usually partnered with oxygen or nitrogen.
|
|
|
In biology, which is stronger- covalent bonds or ionic bonds?
|
Covalent. Ionic bonds break apart in water, which is plentiful in organisms.
|
|
|
Van der Waals forces
|
weak; occur only when atoms and molecules are very close together. Because electrons are in constant motion, they are not always symmetrically distributed in the molecule, resulting in ever-changing "hot-spots" or positive and negative charge that allow atoms and molecules to stick to one another.
|
|
|
Tetrahedron
|
A single "s" and three "p" orbitals; CH4
|
|
|
Molecular structure of water
|
V-shaped, angle of 104.5 degrees
|
|
|
Chemical equilibrium
|
The point at which forward and reverse reactions occur at the same rate. This is a dynamic equilibrium- both the forward and reverse reactions continue to occur, but the concentrations stay the same.
|
|
|
cohesion
|
The binding together of like molecules, often by hydrogen bonds.
|
|
|
adhesion
|
The binding together of unlike molecules; the clinging of one substance to another.
|
|
|
surface tension
|
Related to cohesion; a measure of how difficult it is to stretch or break the surface of a liquid.
|
|
|
Water has a _____ specific heat
|
Water has a high specific heat. Because of this, water will change its temperature less when it absorbs or loses a given amount of heat.
|
|
|
specific heat
|
The amount of heat that must be absorbed or lost for one gram of that substance to change its temperature by 1 degree celsius.
|
|
|
heat of vaporization
|
The quantity of heat a liquid must absorb for one gram of it to be converted from the liquid to the gaseous state.
|
|
|
Water has a ____ heat of vaporization.
|
Water has a high heat of vaporization.
|
|
|
evaporative cooling
|
As a liquid evaporates, the surface of the liquid that remains behind cools down. This occurs because the "hottest" molecules, those with the greatest kinetic energy, leave as a gas, taking heat with them.
|
|
|
At what temperature does water reach its greatest density?
|
4 degrees celsius
|
|
|
solution
|
A liquid that is a completely homogeneous mixture of two or more substances.
|
|
|
aqueous solution
|
Water is the solvent
|
|
|
hydration shell
|
Water surrounds the individual dissolved ions in a solution. The positively-charged hydrogens surround anions, while the negatively-charged oxygens surround cations.
|
|
|
mole
|
Equal in number to the molecular weight of the substance. 6.022x10^23
|
|
|
molarity
|
The number of moles of solute per liter of solution.
|
|
|
molecular weight
|
The sum of all the weights of all the atoms in a molecule; thus, the molecular weight of sucrose (C12H22O11) is 342 daltons (12*12 + 22*1 + 11*16).
|
|
|
hydroxide ion
|
-OH
Charge of -1 |
|
|
hydronium ion
|
H3O+
Charge of +1 |
|
|
acid
|
A substance that increases the H+ concentration of a solution.
|
|
|
base
|
A substance that reduces the hydrogen ion concentration in a solution.
|
|
|
The product of the concentrations of H+ and OH-=
|
[H+][OH-] = 10^-14 M
|
|
|
pH=
|
-log[H+]
|
|
|
buffers
|
Substances that minimize changes in the concentrations of H+ and OH- in a solution.
|
|
|
pH of human blood
|
7.4
|
|
|
Common biological buffer
|
Carbonic acid (H2CO3) dissociates to yield a bicarbonate ion (HCO3-) and a hydrogen ion (H+).
|
|
|
acid precipitation
|
Refers to rain, snow, or fog more acidic than pH 5.6
|
|
|
organic chemistry
|
The branch of chemistry that specializes in the study of carbon compounds.
|
|
|
hydrocarbons
|
Organic molecules consisting only of carbon and hydrogen.
|
|
|
isomers
|
Compounds that have the same molecular formula but different structures and hence different properties.
|
|
|
geometric isomers
|
Have all the same covalent partnerships, but they differ in their spatial arrangements.
|
|
|
structural isomers
|
Differ in the covalent relationships of their atoms.
|
|
|
enantiomers
|
Mirror images of each other. Two enantiomers of a drug (R, S) may have different properties.
|
|
|
functional groups
|
The components of organic molecules that are most commonly involved in chemical reactions. A molecule's properties are inherent in its functional groups.
|
|
|
hydroxyl group
|
-OH. Alcohols. Makes the whole molecule polar.
|
|
|
carbonyl group
|
C=O. Ketone if in middle, aldehyde if on end. Carbs?
|
|
|
carboxyl group
|
Carbonyl + hydroxyl. Strong acid, exists as an ion.
|
|
|
amino group
|
-NH2. Acts as a base because it can pick up a proton.
|
|
|
sulfhydryl group
|
-SH. Thiols. Disulfide bonds- molecular fastener.
|
|
|
phosphate groups
|
PO4. Unstable, negatively charged.
|
|
|
adenosine triphosphate
|
Primary energy-transferring molecule in the cell.
|
|
|
amino acid structure
|
Carbon bonded to an H, an amino group, a carboxyl group, and an R group.
|
|
|
What happens to the charge of an amino acid at physiological pH?
|
The carboxyl group loses an H and the amino group gains an H, so the amino acid becomes charged.
|
|
|
Side chains with ring structures
|
Phe, Tyr, Trp. Bulky, not used very often, mostly used for structural support.
|
|
|
Definition of protein
|
A polymer of amino acids linked to one another by peptide bonds.
|
|
|
peptide bonds
|
Bond between the carbonyl carbon and amino nitrogen.
|
|
|
How is the primary structure of a protein written?
|
From the N-terminus to the C-terminus.
|
|
|
primary structure
|
A protein's unique sequence of amino acids.
|
|
|
secondary structure
|
Result of hydrogen bonds at regular intervals along the polypeptide backbone. Alpha helix, beta pleated sheet.
|
|
|
tertiary structure
|
Consists of irregular contortions from bonding between side chains- hydrogen bonds, ionic bonds, hydrophobic interactions, and Van der Waals forces.
|
|
|
disulfide bridges
|
Covalent bonds that reinforce the protein's conformation. Form between two cysteine monomers.
|
|
|
Sickle Cell Disease
|
Single mutation in hemoglobin. Glutamic acid is replaced by valine, which is hydrophobic.
|
|
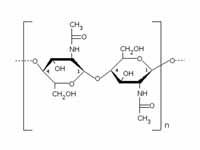
What is this structure?
|
chitin
|
|
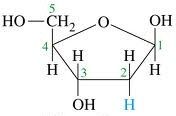
What is this structure?
|
deoxyribose
|
|
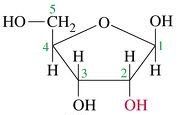
What is this structure?
|
ribose
|
|
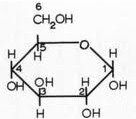
What is this structure?
|
alpha glucose
|
|
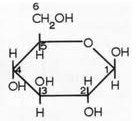
What is this structure?
|
beta glucose
|
|
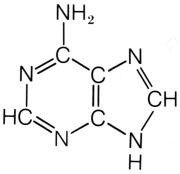
What is this structure?
|
adenine
|
|
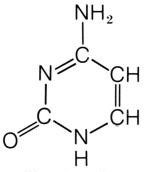
What is this structure?
|
cytosine
|
|
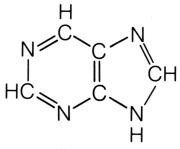
What is this structure?
|
purine
|
|
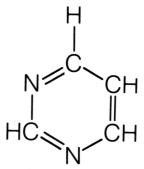
What is this structure?
|
pyrimidine
|
|
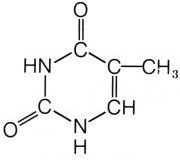
What is this structure?
|
thymine
|
|
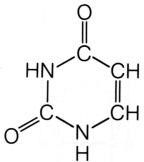
What is this structure?
|
uracil
|
|
|
Two values to consider in microscopy
|
Magnification and resolving power
|
|
|
Resolving Power
|
AKA resolution
Measure of the clarity of the image; the minimum distance two points can be separated and still be distinguished as two separate points. |
|
|
Best resolution of light microscopes
|
.2 micrometers
This resolution is limited by the wavelength of the visible light used to illuminate the specimen. |
|
|
Electron Microscopes
|
Focus a beam of electrons through the specimen. The practical resolution limit for biologists is about 2 nm, although it can achieve about .1 nm.
A disadvantage is that the specimen must be nonliving or dead. |
|
|
Cell ultrastructure
|
Term biologists use to refer to a cell's anatomy as resolved by an electron microscope.
|
|
|
Transmission electron microscope
|
Aims an electron beam through a thin section of the specimen. Uses electromagnets as lenses to focus and magnify the image by bending the paths of the electrons. Used mainly to study the internal ultrastructure of cells.
|
|
|
Scanning electron microscope
|
Useful for detailed study of the surface of the specimen.
The electron beam scans the surface of the sample, which is usually covered with a thin film of gold. |
|
|
Cytology
|
The study of cell structure
|
|
|
Artifacts (microscopy)
|
Structural features seen in microscopy that do not exist in the living cell.
|
|
|
Cell fractionation
|
Takes cells apart, separating the major organelles so that their individual functions can be studied. Uses a centrifuge to fractionate cells.
|
|
|
Ultracentrifuges
|
Powerful machines the can spin as fast as 80,000 rpm and apply forces on particles of up to 500,000 times the force of gravity.
|
|
|
Homogenization
|
Step of fractionation- the disruption of cells.
The objective is to break the cells apart without severely damaging their organelles. Spinning the homogenate in a centrifuge separates the parts of the cell into two fractions: the pellet and the supernatant. |
|
|
Pellet
|
The larger structures of a cell that, when centrifuged, become packed at the bottom of the test tube.
|
|
|
Supernatant
|
The smaller parts of the cell suspended in liquid above the pellet when centrifuged. The supernatant is decanted into another test tube and centrifuged again.
|
|
|
Prokaryotic Cells
|
Only bacteria and archaea.
Has no nucleus. Genetic material is concentrated in a region called the nucleoid (no membrane). |
|
|
Eukaryotic Cells
|
Protists, plants, fungi, and animals.
Has a true nucleus enclosed by a nuclear membrane. |
|
|
Cytoplasm
|
The entire region between the nucleus and the membrane bounding the cell. Consists of the cytosol and organelles.
|
|
|
Cytosol
|
Semifluid medium in which organelles are suspended.
|
|
|
Mycoplasmas
|
The smallest cells known.
Bacteria which have diameters between .1 and 1.0 micrometer. |
|
|
Eukaryotic cell size
|
Typically 10 to 100 micrometers in diameter.
|
|
|
Bacteria cell size
|
1 to 10 micrometers in diameter.
|
|
|
Ratio of surface area to volume
|
The smaller the object, the greater its ratio of surface area to volume.
Volume grows more than surface area. |
|
|
Nucleus
|
Contains most of the genes that control the eukaryotic cell.
Averages about 5 micrometers in diameter. The nuclear envelope encloses the nucleus. |
|
|
Where are genes located besides the nucleus?
|
Some genes are located in the mitochondria and chloroplasts.
|
|
|
Nuclear envelope
|
Double membrane; each membrane is a lipid bilayer.
The two membranes are separated by a space of about 20 to 40 nanometers. Perforated by pores and pore complexes. |
|
|
Pore Complex
|
Lines each pore and regulates the entry and exit of certain large macromolecules and particles.
|
|
|
Nuclear lamina
|
The nuclear side of the nuclear envelope.
A netlike array of protein filaments that maintains the shape of the nucleus. |
|
|
chromatin
|
DNA is organized into a diffuse mass called chromatin.
|
|
|
chromosomes
|
As the cell prepares to divide the chromatin coils up into chromosomes.
|
|
|
Nucleolus
|
Prominent structure in the nondividing nucleus.
Components of ribosomes are synthesized and assembled here. These components then pass through the nuclear pores and are assembled into ribosomes in the cytoplasm. |
|
|
mRNA
|
messenger RNA.
Synthesized in the nucleus. Conveys genetic messages to the cytoplasm via the nuclear pores. Once in the cytoplasm, the mRNA attaches to ribosomes, where the genetic message is translated into the primary structure of a specific protein. |
|
|
Ribosomes
|
The sites where the cell makes proteins.
|
|
|
Free ribosomes
|
Suspended in the cytosol.
Most of the proteins made by free ribosomes will function within the cytosol. |
|
|
Bound ribosomes
|
Attached to the outside of the endoplasmic reticulum.
Generally make proteins that are destined either for inclusion into membranes or for export from the cell. |
|
|
endomembrane system
|
Includes the nuclear envelope, ER, Golgi apparatus, lysosomes, various kinds of vacuoles, and the plasma membrane.
|
|
|
endoplasmic reticulum
|
Accounts for more than half the total membrane in many eukaryotic cells.
Consists of a network of membranous tubules and sacs called cisternae. |
|
|
Smooth ER
|
Cytoplasmic surface lacks ribosomes.
Functions in diverse metabolic processes, including synthesis of lipids, metabolism of carbohydrates, and detoxification of drugs and poisons. |
|
|
Rough ER
|
Ribosomes stud the cytoplasmic surface of the membrane.
Produce proteins for secretion. |
|
|
Glycoproteins
|
Most secretory proteins.
Proteins that are covalently bonded to carbohydrates. The carbohydrate appendage of a glycoprotein is called an oligosaccharide. |
|
|
Transport Vesicles
|
Transport secretory proteins from the rough ER to the plasma membrane.
Develop in the transitional ER. |
|
|
Golgi apparatus
|
Transport vesicles travel from the ER to Golgi.
The products of the ER are modified and stored, and then sent to other destinations. |
|
|
Lysosome
|
A membrane-bounded sac of hydrolytic enzymes that the cell uses to digest macromolecules.
Enzymes work best in an acidic environment, about pH 5. |
|
|
Phagocytosis
|
Protists and macrophages engulf smaller organisms or food particles.
|
|
|
Autophagy
|
Lysosomes use their hydrolytic enzymes to recycle the cell's own organic material.
|
|
|
Pompe's Disease
|
The liver is damaged by an accumulation of glycogen due to the absence of a lysosomal enzyme needed to break down the polysaccharide.
|
|
|
Tay-Sachs Disease
|
A lipid-digesting enzyme is missing or inactive, and the brain becomes impaired by an accumulation of lipids in the cells.
|
|
|
Contractile vacuoles
|
Pump excess water out of the cell.
Found in many freshwater protists. |
|
|
Food vacuoles
|
Formed by phagocytosis.
|
|
|
Vacuoles
|
Membrane-bounded sacs within the cell.
Larger than vesicles. |
|
|
Central vacuole
|
Large organelle in plant cells enclosed by a membrane called the tonoplast. Versatile storage.
|
|
|
Gram Stain
|
Distinguishes between two different kinds of bacterial cell walls (cell wall composition).
Bacteria are stained with a violet dye and iodine, rinsed in alcohol, then stained again with a red dye. The structure of the cell wall determines the staining response. |
|
|
Gram-Positive Bacteria
|
Have cell walls with a large amount of peptidoglycan that traps the violet dye.
Have simpler cell walls. Generally less likely to cause disease. |
|
|
Gram-Negative Bacteria
|
Contains less peptidoglycan. The violet dye is easily rinsed away, but the red dye remains.
An outer membrane on the cell wall contains lipopolysaccharides, carbs- can be toxic. More likely to be antibiotic resistant, disease-causing. |
|
|
peptidoglycan
|
Found in most bacterial cell walls.
A network of sugar polymers cross-linked by polypeptides. The effect is a single molecular network enclosing and protecting the entire cell (strength and elasticity). |
|
|
capsule (bacteria)
|
Many prokaryotes secrete a sticky, jelly-like coating of lipopolysaccharides that forms another protective layer.
|
|
|
penicillin
|
Inactivates the enzyme that synthesizes cross-links in peptidoglycan, preventing the formation of a functional cell wall.
Esp. useful in gram-positive bacteria. |
|
|
chaperonins
|
Protein molecules that assist the proper folding of other proteins.
|
|
|
aminoacyl-tRNA synthetase
|
Joins each amino acid to the correct tRNA.
There are 20 of these enzymes in the cell, one for each AA. |
|
|
NAD+
|
Coenzyme.
During respiration, hydrogen atoms are stripped from glucose and transferred to NAD+. Functions as an oxidizing agent. Accepts an electron and becomes NADH. |
|
|
NADH
|
Reduced form of NAD+.
Electrically neutral. Electrons fall down the ETC from NADH to oxygen. |
|
|
phosphorylation
|
Primes a molecule to undergo some kind of change that performs work, and the molecule loses its phosphate group in the process.
|
|
|
FAD
|
An electron acceptor similar to NAD+.
FADH2 is its reduced form. |
|
|
1 molecule of glucose makes approximately how much ATP?
|
Approx. 38 molecules of ATP
|
|
|
Oxidative vs. substrate-level phosphorylation
|
Oxidative is responsible for 90% of ATP production, while substrate-level is responsible for 10% of ATP production.
|
|
|
glycolysis
|
A series of ten chemical reactions.
Glucose is broken down into two 3-carbon molecules of pyruvate. Occurs in the cytoplasm. |
|
|
acetyl CoA
|
Pyruvate enters the mitochondrion. It is then converted into acetyl CoA.
Acetyl CoA enters the Krebs cycle. |
|
|
ATP synthase
|
Protein complex.
Found in the inner membrane of the mitochondrion. Enzyme that makes ATP. Turbine that uses the flow of H+ to power ATP production. Ion pump running in reverse. H+ is produced in the ETC, and creates a concentration gradient that ATP synthase takes advantage of. |
|
|
chemiosmosis
|
Step of respiration after the ETC.
Uses ATP synthase to produce ATP from the products of the ETC. |
|
|
Fermentation
|
Method of generating ATP in anaerobic conditions.
Much less ATP than oxidative phosphorylation. Consists of glycolysis plus reactions that regenerate NAD+, which can be reused by glycolysis. |
|
|
obligate anaerobes
|
Carry out fermentation or anaerobic respiration and cannot survive in the presence of O2.
|
|
|
facultative anaerobes
|
Can use either fermentation or cellular respiration.
Pyruvate is a fork in the metabolic road that leads to two alternative catabolic routes. Yeast |
|
|
insulin
|
peptide hormone.
Produced by the pancreas. Production is stimulated by hyperglycemia, or an increase in blood sugar. Promotes transport of glucose into body cells and stimulates liver and muscle cells to store glucose as glycogen. |
|
|
glucagon
|
Peptide hormone that is synthesized by the pancreas.
Opposite to insulin. Primary function is to increase levels of glucose in the blood. |
|
|
lipolysis
|
When glycogen stores are depleted, glucagon triggers lipolysis, which converts fat (stored in adipose cells) to glycerol and fatty acids.
|
|
|
adipose cells
|
Store fat.
|
|
|
glycogen
|
A polymer of glucose, extensively branched.
Animals store energy in the form of this polysaccharide. |
|
|
Advanced Glycation End-Products
|
A sugar molecule bonds to either a protein or a lipid molecule without an enzyme to control the reaction.
Similar to glycosylation. |
|
|
Glycosylation
|
A chemical reaction in which glycans are attached to proteins, lipids, or other molecules, forming glycoproteins.
|
|
|
Four properties of water
|
-cohesive behavior
-ability to moderate temperature -expansion upon freezing -versatility as a solvent |
|
|
steroids
|
Lipids characterized by a carbon skeleton consisting of four fused rings.
|
|
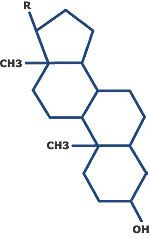
What is this?
|
Steroid. Notice the four fused rings.
|
|
|
Rosalind Franklin
|
First predicted the helical structure of DNA using x-ray crystallography.
|
|
|
Chargraff's Rules
|
Predicted DNA base pairing.
Adenine=Thymine Guanine=Cytosine Adenine+Guanine=Thymine+Cytosine |
|
|
amylase
|
Breaks down starch.
Digestive enzyme |
|
|
pepsin
|
Digestive enzyme that breaks down proteins.
|
|
|
lipase
|
Digestive enzyme that breaks down fat.
|
|
|
lactase
|
Breaks down lactose.
|
|
|
Robert Hooke
|
1665- coined the term cell while studying thin slices of cork.
|
|
|
Anton van Leeuwenhoek
|
Discovered single-celled organisms using handcrafted microscopes in 1673.
Discovered a microbial world in droplets of pond water. |
|
|
Schleiden and Schwann
|
In 1839, these two German biologists acknowledged cells as the ubiquitous units of life.
"All living things consist of cells" |
|
|
cell fractionation
|
Takes cells apart and separates the major organelles from one another, by size, using centrifuges.
|
|
|
freeze-fracture microscopy
|
Cells are frozen and then a knife is used to crack them open. The crack often passes through the two layers of the plasma membrane.
|
|
|
transmission electron microscopy (TEM)
|
A beam of electrons is sent through a thin specimen. Allows for imaging of the interior of cells.
|
|
|
scanning electron microscopy (SEM)
|
Directs electrons to the surface of the specimen. The 3D surface of the object can be seen.
|
|
|
nucleolus
|
A region in the nucleus active in the synthesis of ribosomal RNA and ribosome assembly.
|
|
|
nuclear envelope
|
The double membrane in eukaryotes that encloses the nucleus, separating it from the cytoplasm.
|
|
|
nuclear lamina
|
The nuclear side of the nuclear envelope containing protein filaments that maintain the shape of the nucleus.
|
|
|
nuclear pore complex
|
The multiprotein structure forming a channel through the nuclear envelope, allowing selected molecules (proteins, RNA, ribosomes) to move between the nucleus and the cytoplasm.
|
|
|
endoplasmic reticulum
|
A network of membranous tubules and sacs within the cytoplasm of eukaryotic cells where lipids are synthesized and membrane-bound proteins and secretory proteins are made.
|
|
|
microtubules
|
25 nm in diameter.
Found in the cytoplasm of all eukaryotic cells. Constructed from a globular protein called tubulin. Shape and support the cell. Serve as tracks along with organelles containing motor molecules can move. Involved in the separation of chromosomes during cell division. |
|
|
microfilaments
|
AKA actin filaments
~7 nm in diameter Built from molecules of actin, a globular protein. Seem to be present in all eukaryotic cells. Structural role in the cytoskeleton is to bear tension (pulling forces). Part of the contractile apparatus of muscle cells (paired with myosin). |
|
|
dynein
|
large motor protein
Allows one fiber to "walk" along another. Process requires ATP. |
|
|
kinesin
|
Motor protein similar to dynein.
Move along microtubules, and are powered by ATP. Supports cellular functions such as mitosis and meiosis. |
|
|
cilia
|
Movement controlled by microtubules.
Occur in large numbers on a cell surface. Anchored to the cell by a basal body. Work like oars to generate a force in a direction perpendicular to the cilium's axis. Circulate fluids, move egg into oviduct, line air passages. |
|
|
basal body
|
Structurally identical to a centriole.
Anchors cilia and flagella to cells. |
|
|
flagella
|
Usually limited to one or two per cell.
Anchored to the cell by a basal body. Undulating motion generates force in the same direction as the flagellum's axis. Required for sperm motility. |
|
|
cytoplasmic streaming
|
A circular flow of cytoplasm within cells.
Speeds distribution of materials. In plant cells, actin-myosin interactions drive streaming. |
|
|
pseudopodia
|
Localized contraction caused by actin and myosin play a role in ameboid movement. Pseudopodia are cellular extensions that extend and contract through the reversible assembly of actin subunits into networks that convert cytoplasm from sol to gel.
|
|
|
sol vs. gel
|
Cytoplasm:
Streaming, or fluid, cytoplasm is known as sol. Nonmoving cytoplasm is known as gel. |
|
|
intermediate filaments
|
8 to 12 nm in diameter. Between the size of microtubules and microfilaments.
Specialized for bearing tension. More permanent fixtures of cells than filaments and tubules. Important in reinforcing the shape of a cell and fixing the position of certain organelles. |
|
|
plasmodesmata
|
Channels that perforate the walls of plant cells.
Cytoplasm passes through these channels and connects the living contents of adjacent cells. The membrane lines the channel. |
|
|
tight junctions
|
Animal cells.
Membranes of neighboring cells are pressed together, preventing leaking of extracellular fluid. |
|
|
desmosomes
|
Animal cells.
Anchoring junctions Fasten cells together into strong sheets. Use intermediate filaments. |
|
|
gap junctions
|
Animal cells.
Communicating junctions Provide cytoplasmic channels between adjacent cells. |
|
|
central dogma
|
Summarizes the flow of information in cells.
DNA-->RNA-->proteins |
|
|
transcription
|
DNA is transcribed to mRNA.
Synthesis of RNA under the direction of DNA. Takes place in the nucleus. |
|
|
translation
|
mRNA is translated to proteins.
Actual synthesis of a polypeptide, which occurs under the direction of mRNA. |
|
|
carotenoids
|
Similar to chlorophyll.
Meant to protect chlorophyll from photodamage. |
|
|
Calvin Cycle
|
Dark reaction of photosynthesis.
Requires CO2. Occurs in the stroma. |
|
|
G3P
|
Molecules produced by the Calvin Cycle.
Often used to make glucose and fructose. In rapidly photosynthesizing cells where there is an abundance of sucrose, glucose is temporarily stored in the chloroplast as starch. The starch is then broken down at night and used to make more sucrose. |
|
|
integrins
|
Allow for communication between the cell and the ECM.
Transmembrane proteins which bind to ligands found in the ECM. |
|
|
fibronectins
|
Glycoproteins in the ECM that bind to integrins and play a major role in cell adhesion, growth, migration, and differentiation.
|
|
|
cohesins
|
Proteins that regulate the separation of sister chromatids during cell division.
|
|
|
osmosis
|
The movement of water from low solute concentration to high solute concentration.
|
|
|
alcohol fermentation
|
Pyruvate is converted into ethanol in two steps.
The first step releases CO2 from the pyruvate, which becomes acetaldehyde. In the second step, acetaldehyde is reduced by NADH to ethanol. This regenerates the supply of NAD+ needed for glycolysis. Pyruvate, NADH-->ethanol, CO2, NAD+ |
|
|
lactic acid fermentation
|
Pyruvate is reduced directly by NADH to form lactate as a waste product, with no release of CO2.
|
|
|
NAD+ is converted to NADH when it gains...
|
2 electrons and 1 proton
|
|
|
Phosphofructokinase
|
Step 3 of glycolysis.
Allosteric enzyme that phosphorylates fructose 6-phosphate. |
|
|
allosteric enzyme
|
Enzymes that change their conformation upon binding to certain noncompetitive inhibitors.
|
|
|
transcription
|
The synthesis of RNA under the direction of DNA.
|
|
|
Translation
|
The actual synthesis of a polypeptide, which occurs under the direction of mRNA.
|
|
|
central dogma
|
DNA-->RNA-->proteins
|
|
|
template strand
|
Strand of DNA that is transcribed by RNA. The other exists only to help replicate DNA. Can switch off between strands.
|
|
|
promoter
|
Region of DNA where RNA polymerase attaches and initiates transcription.
|
|
|
RNA polymerase
|
Pries the two strands of DNA apart and hooks together the RNA nucleotides as they base-pair along the DNA template.
Can add nucleotides only to the 3' end of the growing polymer. |
|
|
DNA polymerase
|
Functions in DNA replication.
Can add nucleotides only to the 3' end of the growing polymer. |
|
|
RNA Splicing
|
The sequence of DNA nucleotides that codes for a polypeptide is not continuous. The noncoding segments of nucleic acid that lie between coding regions are called introns. The parts that are eventually translated into amino acid sequences are called exons.
Removal of a large portion of the RNA molecule that is initially synthesized into just the parts needed for the protein. |
|
|
introns
|
Noncoding regions taken out during RNA splicing.
"Intervening sequences" |
|
|
exons
|
Regions of RNA transcripts that are expressed as amino acid sequences.
Survive the splicing process. "Expressed sequences" |
|
|
spliceosomes
|
Proteins that carry out RNA splicing. Within the spliceosome, base pairing between the pre-mRNA and snRNA promotes cleavage and ligation.
|
|
|
snRNPs
|
Small nuclear ribonucleoproteins.
There are short nucleotide sequences at the end of introns, and snRNPs recognize these splice sites. Located in the cell nucleus and composed of RNA and protein molecules. |
|
|
snRNA
|
RNA in a snRNP particle.
Small nuclear RNA. Each molecule is about 150 nucleotides long. |
|
|
ribozyme
|
An enzymatic RNA molecules that catalyzes reactions during RNA splicing.
|
|
|
Function of introns
|
Introns play regulatory roles in the cell.
Some genes are known to give rise to two or more different proteins, depending on which sequences are treated as exons during RNA splicing. |
|
|
domains
|
Many proteins have a modular architecture consisting of discrete structural and functional components called domains. One domain might include the active site, which another might attach the protein to a cellular membrane.
|
|
|
tRNA
|
Transfer RNA.
The function of tRNA is to transfer amino acids from the cytoplasm to a ribosome. Each type of tRNA molecule links a particular mRNA codon with a particular amino acid. |
|
|
anticodon
|
One end of the tRNA that binds to a complementary codon on the mRNA sequence in a ribosome.
|
|
|
wobble
|
A violation of the base-pairing rules in the third nucleotide (5' end) of a tRNA anticodon, allowing it to form hydrogen bonds with more than one kind of base in the third position (3' end) of a codon.
|
|
|
aminoacyl tRNA-synthetase
|
Enzyme that joins a specific amino acid to the correct tRNA.
There are 20 of these enzymes in the cell, one for each amino acid. |
|
|
APE- ribosomes
|
A site- holds the tRNA carrying the next amino acid to be added to the chain.
P site- holds the tRNA carrying the growing polypeptide chain. E site- exit site. |
|
|
Stages of building a polypeptide
|
Initiation- brings together mRNA, a tRNA bearing the first amino acid of the polypeptide, and the two subunits of a ribosome.
Elongation- amino acids are added one by one to the first amino acid. Termination- a stop codon reaches the A site of the ribosome. A release factor frees the polypeptide from the ribosome. |
|
|
release factor
|
A protein that binds directly to the stop codon of an mRNA strand when it reaches the A site in a ribosome. Causes the addition of a water molecule instead of an amino acid to the polypeptide chain. This reaction hydrolyzes the completed polypeptide from the tRNA that is in the P site, freeing the polypeptide from the ribosome.
|
|
|
GTP
|
guanosine triphosphate
A molecule closely related to ATP. Provides energy for chain initiation and elongation in polypeptide synthesis. |
|
|
base-pair substitution
|
The replacement of one nucleotide and its partner in the complementary DNA strand with another pair of nucleotides.
They have no effect on the encoded protein. |
|
|
insertions and deletion
|
Addition or subtraction of one or more nucleotide pairs in a gene.
Have a disastrous effect on the resulting protein. May alter the reading frame, producing a frameshift mutation. |
|
|
Frame-shift mutations
|
Single bases inserted or deleted- usually leads to nonfunctional proteins.
|
|
|
Silent mutations
|
A substitution mutation.
Do not change the amino acid specified by the codon. |
|
|
Missense mutation
|
Type of substitution mutation.
Code for the wrong amino acid. |
|
|
Nonsense mutation
|
Type of substitution mutation.
Code for a stop codon instead of an amino acid codon, nearly always leading to a nonfunctional protein. |
|
|
spontaneous mutation
|
Can occur during DNA replication, recombination, or repair.
|
|
|
mutagens
|
Physical or chemical agents that can cause mutations.
|
|
|
Ames test
|
A method of testing the mutagenic activity of different chemicals.
Uses easily-grown bacteria as test organisms. |
|
|
operon
|
Model of gene expression.
Several genes of related function are grouped into one transcription unit that has a single "on-off switch" to control the entire cluster. Contains genes, the promoter, and an operator. |
|
|
operator
|
"On-off switch" in an operon.
Lies between the promoter and the first gene. |
|
|
promoter
|
Where RNA polymerase binds.
Operons- a single promoter regulates the synthesis of one polycistronic mRNA. |

