![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
92 Cards in this Set
- Front
- Back
|
Endonuclease |
2 aspartate residues coordinate two Mg(2+) ions Occurs in the palm domain ‘Two metal ion mechanism’ |
|
|
Role of aspartic acid in endonuclease |
(-) of aspartic acid allows it to hold onto 2 divalent metal ions, Mg(2+) Mg(2+) positioned to catalyze certain reactions |
|
|
Metal A in endonuclease |
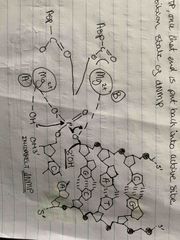
Activates 3’OH on primer 3’OH nucleophilic attack on alpha-phosphate of new dNTP Mg(2+) converts 3’OH to 3O-, by dissociating protons Extra e- pair forms phosphodiester bond & breaks phosphoenhydryde bond To do this (-) a.a sidechain holds triphosphate dNTP in place
|
|
|
Metal B in endonuclease |
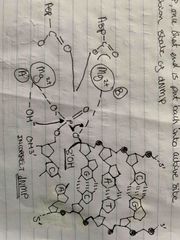
Plays dual role Stabilizes (-) charge building up on leaving Oxygen Holds beta and gamma phosphate in place Products= pyrophosphate triggering translocation of the polymerase to post insertion site Fast reaction does not require ATP |
|
|
Exonuclease |
Also has aspartate but uses different nucleophile Similar to two metal ion mechanism |
|
|
Metal A exonuclease |
Facilitates formation of attacking OH ion Which attacks phosphodiester bonds of last dNTP du to WATER IN ACTIVE SITE Metal a lowered pKA of water to form OH (-) Unpaired e(-) pair attacks terminal P of DNTP and breaks phosphodiester bonds to release that dNMP
|
|
|
Metal B exonuclease |
Helps leaving 3’OH left after terminal nucleotide is removed on remaining DNA 3’OH needed to be a nucleophile for addition of correct dNTP Stabilized transition site of dNMP |
|
|
Processsivity |
Ability to catalyze consecutive reactions without releasing substrate DNA pol = low processivity |
|
|
Fidelity |
Accuracy in replicating from template Some have large range |
|
|
E. Coli has 5 pol, describe 2 important ones |
Pol1: used to link Okazaki fragments Pol3: main enzyme in chromosome replication can correct its own mistakes from addition of nucleotides due to 3’-5’ exonuclease Unlike pol1, it does not contain 5’-3’ exonuclease activity |
|
|
Nick translation, pol1 |
5’-3’ exo.N activity same as DNA synthesis unique to pol1 Removes RNA primer between Okazaki fragments Removes dNMP’s (rna n) and adds dNTP’s (Dna n) Rxn involves Mg(2+) dependent mechanism |
|
|
Initiation |
1st step of E. coli rep Primary control over rep is exerted Process begins at origins Initiation factors (proteins) use these sites |
|
|
Elongation |
Leading strand synthesis Rna primers bind and direct beta clamp & assembly of leading strand pol3 core complexes Core c’s^ advance rep to opposite DnaB Dna pol3 added to clamp Polymerase will collide w dnab & follows it as it separates strands (polymerase is activated) |
|
|
Activated polymerase in elongation |
Binds to large section of SSDNA Dna has primers +d by primase Starts to make lagging strand |
|
|
Elongation by replisome |
Replisome is multi-enzyme complex works cooperatively and quickly to copy leading and lagging strand 1 complex at each replication fork |
|
|
Termination |
Involves specific sites of proteins too |
|
|
Origins |
DNA sequences Binding site for initiator protein that orchestrates assembly of replication initiation machinery Stretch of A-T rich DNA that unwinds readily but not spontaneously |
|
|
Directions of DNA sequences |
13-Mer site & 9-mer site Recognized by specific proteins Ie) DnaA rep initiator protein |
|
|
DnaA |
Wraps DNA around oligomer Bonds with HU which allows conformational change to the open complex formation |
|
|
HU |
Similar to chromatin folding & DNA packaging proteins Once bound to DNA and has ATP at the active site, open complex formation is induced |
|
|
Open complex |
Consist of: origin of rep, DnaA, ATP and HU Partial helix straining nearest DNA complex, which is 13-mer repeat section Induces strand separation |
|
|
Induced strand separation |
Caused by open complex DnaB is helped by DnaC DnaC is a protein that binds to ATP & has an affinity for DnaB This affinity causes ring to open up to be loaded onto rep fork Each will move in opposite directions expanding s.s DNA |
|
|
Assembly of e.coli replication forks |
DnaB expand rep fork bubble Primase forms rna primer at each fork Fork movement of DnaB will dislodge DnaA Primase adds first nucleotides to become leading strand and rna primer Rna primer on either end Supercoil stress in dna must be removed by the topoisomerase enzyme |
|
|
A=T rich 13-mer repeats |
to one side of DnaA 9 mer sites 3 A=T rich direct repeats of 13 bp repears are DNA unwinding element unwind readily after binding of the initiator |
|
|
DnaA 9-mer sites |
4 copies of a nine-nucleotide consensus sequence to which bacterial initiator protein DnaA binds called R sites for 'repeat' |
|
|
DnaA |
Initiator protein. at E. coli oriC member AAA+ protein (conformational changer) Wraps DNA around oligomer Bonds with HU which allows conformational change to the open complex formation |
|
|
Activation of the replication Origin |
Once DNA is in the open complex and ss regions are exposed Two hexamers of the DnaB helicase assemble DnaC is required for loading dnaB on ssDNA open complex |
|
|
DnaC |
member of AAA+ protein family pries open the hexameric ring of dnaB and slips onto ssDNA bubble |
|
|
Prepriming complex |
a complex of proteins assembled at oriC in e.coli at an early stage of the replication fork assembly |
|
|
activation. of the E.coli oriC |
1) DnaA oligomerizes on. binding the origin & wraps dna around oligomer. HU helps open complex formation 2) DnaC loads dnaB onto each strand & forms pre-priming complex 3) dnaB expands rep. bubble & primase forms RNA primers at each rep. fork 4) rna primers direct clamp loading & assembly of of a PolIII holoenzyme complex , which advances rep to opposite dnaB 5) Continies dnaB activity allows lagging-s priming & Okazaki fragment synthesis |
|
|
Assembly of bacterial replication |
1) ATP on dnaB allows it to. translocate and unwind DNA, dislodging the dnaA protein. 2) unwinding generates supercoil stress in the DNA ahead of the replication fork and this stress must be removed by topoisomerase 3) newly unwound DNA is coated with single-stranded bring proteins to protect it 4) RNA primers are synthesized by primase |
|
|
ARS |
In euk initiation ARS (auto rep sequences) 100-200 bp long contains conserved A seq. for initiator binding & B1 B2 AND B3 elements (AT-rich) |
|
|
origin recognition complex (ORC) |
In euk initiation subunits w actions similar to dnaA ATP is required for orc binding to the origin |
|
|
Beta clamp |
homodimers, shaped like a ring that encircles the DNA duplex holds the polymerase onto the DNA while sliding along the duplex converts the PolII core from a distributive enzyme to one that stays attached to dna during repeptitive cycles of dNMP incorporation assist processivity |
|
|
Pol III core |
similar to Pol 1 structure with the palm, thumb and finger domains alpha subunit is the main replicative subunit, recruits a subunit for 3'-5' proofreading activity |
|
|
E.coli beta clamp loader |
beta sliding clamp does not assemble onto dna by itself, requires the multiprotein clamp loader to open and close the ring around the DNA clamp loader within the Pol III holoenzyme |
|
|
Clamp loader mechanism |
1) ATP binds to the T subunits, induces a conformational change that enables the clamp loader to bind and open
2) ATP binding is needed for the clamp loader to bind to DNA 3) ATP hydrolysis reverts clamp loader to a form which ejects the clamp loader and therefore allows the clamp to close around dna |
|
|
Trombone model mechanism |
- lagging strand pol finishes o.f - pol detaches from the b clamp & leaves it behind on DNA - pol transfers to a new RNA primer to extend the next fragment - associates w new rna-primed site on which a new b clamp has been assembled |
|
|
1st step Clamp recycling by DNA pol I and ligase |
pol III core dissociates from b clamp b clamp site acts to attract pol1 to remove primer using the. 5'-3' exonuclease activity (nick translation |
|
|
2nd step Clamp recycling by DNA pol I and ligase |
Pol1 dissociates from the DNA, leaving behind an ssdna break B clmap site acts to attract the DNA ligase enzyme ligase enzyme is able to seal the break by forming a phosphodiester bond using the 3'OH and 5'P groups |
|
|
4th step Clamp recycling by DNA pol I and ligase |
the unoccupied B clamp is then opened and unloaded (step 4) by excess & subunits of the clamp loader |
|
|
trombone model meaning |
DNA loops repeatedly gorw and collapse on the lagging strand, allowing the replisome to move towards the replication fork, like the movement of a trombone slide. |
|
|
Ter sites |
involved in termination of chromosome rep 2 clusters of 23bp located 1/2 way around oriC oriented in opposite directions Monomeric Tus protein tighlty binds to a ter site & blocks advance of replication for by stopping dnaB helicase |
|
|
Tus |
binds to Ter site in termination of chromosome replication stands for: termination utilization substance |
|
|
Tus-ter complex |
fork -blocking activity is directional (denoted by arrowheads)
Replicaiton forks are blocked when approaching a Tus-Ter complex from one direction (nonpermissive direction), but not when approaching from the opposite (permissive) direction |
|
|
Tus-Ter system and replication speed |
Equal speed: rep forks of equal speed meet at the same time in the terminus region of the chromosome Unequal speed: meet in the terminus region since replication will be stalled in one direction until it is completed in the. other direction |
|
|
Type II Topoisomerase enzyme |
resolves/ unlinks the catenated daughter chromosomes into 2 separate chromosomes |
|
|
Topoisomerase mechanism of action |
type II topoisomerase protein in prokaryotes is a heterodimer, consisting of 2 cleavage core domains 2 ATPase domains linked by a scaffolding domain enzyme pulles catenated dsDNA into cleavage site and through the use of ATP generates a ds break in the DNA fragment |
|
|
Mechanism of topoisomerase break |
through the use of tyrosine in the catalytic site Tyr gives up its proton, forms a nucleophile attacks phosphate in phosphodiester bond (occurs on both strands) temporarily convalently links the DNA strand to the protein forming. a phosphotyrosyl linkage (5'. adduct) hydrolysis reaction occurs once daughter chromosomes are separated to reform the phosphodiester bond
|
|
|
End problem in euk termination 1st problem |
RNA primer at the extreme end is removed for replacement with DNA there is no 3' terminus for DNA polymerase to extend from ss gap cannot be converted to duplex DNA |
|
|
End problem in euk termination 2nd problem |
genetic information in the gap will be lost next round of replication repeated rounds will cause the ends to progressively shorten until genes near the ends are entriely lost |
|
|
Telomerase |
Carries it's own. template and synthesizes ssDNA telomerase reverse transcriptase (TERT) carries a tightly bound, non-coding telomerase RNA (TR) ~1.5 telomere repeat units which match telomere repeat sequence that it uses as a template to extend the 3' terminus of the telomere beyond what was replicated - the reaction occurs in S. phase |
|
|
Telomerase reaction cycle steps 1-3 |
at 3' terminal end of linear DNA, 3 nucleotides of telomere anneal to 3 RNA nucleotides in telomerase - telomerase extends the 3' end of ssDNA by one telomere repeat - After repeat is added., telomerase separates RNA-DNA hybrid and repositions telomere for. extension of the next 6-mer repear. Telomerase synthesizes many telomere repates in one telomerase binding event. |
|
|
Telomerase reaction cycle steps 4&5 |
telomerase extended 3' ssDNA terminus is converted to duplex dna by priming & polymerization machinery used in chromosome rep -3' terminus of a new telomere still has ssDNA due to same RNA primer-removal problem discussed earlier |
|
|
Aneuploidy |
presence of abnormal number of chromosomes in a cell all cancer cells are aneuploid as a result of mitotic segregation defects or chromosomal non-disjunction |
|
|
Chromosomal translocations |
abnormality caused bu rearangment of parts between non-homologous chromosomes can be direct switing of material between 2 chromosomes or large scale deletions of insertions caused by errors during homologous recombination or ds break repair |
|
|
Double stranded break |
in phosphodiester backbone on both strands of DNA at the same site. usually arrise during replication when rep fork encountes are ss break in the template which then becomes propagated into a dsb in both the template and daughter strands Replication is impossible to continue can alos occur from exposure to UV or gamma radiation usually leads to cell death if not repaired |
|
|
Recombinational DNA repair purpose |
group of recombinantion processes directed at the repair of DNA strand breaks or cross-links repair requires presence of another undamaged homologous dsDNA in diploid cell, 2nd dsDNA is usually the 2nd copy of the chromosome or sister chromatid 2nd dsDNA guides repair process by providing a template for genetic info that otherwise would be lost as a consequence of missing nucleotides at the site of a break |
|
|
Recombinational DNA repair process step 1 |
After DSB broken DNA ends are processed by helicases and nucleases 5' ending strands selectively degraded to create 3' ss extensions or overhands at site of the break ssDNA is coated and protected by ss binding proteins (SSBs) |
|
|
Recombinational DNA repair process step 2 |
3' ss extension invase homologous chromosome in a process catalyzed by recombinase enzymes enzymes replace the SSBs invading strand displaces a strand of intact homologous chromosome and bp w the other structure created sometime referred to as a D-loop |
|
|
Recombinational DNA repair process step 3 |
a second strand invasion takes place, similar to the first |
|
|
Recombinational DNA repair process step 4 |
DNA pol-mediated extension of the invading strands after they are paired lengthens them in a matter that restores any lost information at the site of the break uses the invaded homologous chromosome as the template |
|
|
Completing repair of DSB (step 5) pathway A |
lengthened invading strands can simply be displaced by the action of helicases and then anneal to each other. remaining gaps are filled. by. DNA. polymerases DNA ligases complete the repair by ligating the ends & reforming the 2 dsDNA chromosomes |
|
|
Completing repair of DSB (step 5) pathway B |
Rep is completed by ligating the strands while they are still linked. 4 branched corssover junction w all DNA strands intact such that each branch is a segment of dsDNA called a Holliday intermediate (or junction) formed by the extension of D loops specialized enonucleases called Holliday junction resolvases recognize and cleave the holliday intermediates & has 2 outcomes. |
|
|
Noncrossover |
repair of DSB (step 5) pathway B outcome 1 both intermediates are cleaved at sides labeled X of both. are cleaved at sites labeled Y genetic material between. cleavage sites will be exchanged but chromosomes will not |
|
|
Crossover |
repair of DSB (step 5) pathway B outcome 2 if one junction is cleaved at X sites and other at Y sites such that the repair is now from2. separate chromosomes |
|
|
STEPS OF REPAIRING DS BREAK (crossover) |
1) Broken DNA ends processed-create 3' ss overhangs 2) 3' ss extensions invade homologous chromosome, catalyzed by recombinases 3) replicative extension of the invading strand by Pol III occurs 4) strands are ligated while still linked 5) Holliday junction resolvases recognize & cleave one junction at X site & other at Y site 6) Breaks left behind by the cleavage are sealed by DNA ligase |
|
|
D-loop |
displacement loop causef by strand invasion by the 3' ss extension during DSB repair |
|
|
Homologous recombination |
allows faster genetic adaptation to environment. recombination between 2 DNA molecules of a similar sequence. takes place during meiosis & mitosis in eukaryotes in DSBs in all organisms |
|
|
what segregates at 1st cell division & 2nd cell division |
homologous chromosomes sister chromatids |
|
|
Cohesins |
provide the physical link between sister chromatids |
|
|
Roles of crossing over in meiosis |
1) create a physical link for chromosomal segregation 2) sister chromatids are no longer identical, each set of paired chromatids have been exchanged w homologous chromosome generating genetic diversity |
|
|
Initiation of meiotic recombination pt 1 |
as the cell enters meiosis DSB are intentionally introduced at multiple locations along a chromatid of each pair Breaks are not random, occurs at chromosomal 'hotspots' more frequently than other sites in the genome A protein called Spo11 catalyzes the formation of DSBs |
|
|
Spo11 protein |
found in all eukaryotes acts as a dimer and uses active site of Tyr residue as a nucleophile in transesterification reaction |
|
|
Initiation of meiotic recombination pt 2 |
each subunit of spo11 cleaves a dna strand w phosphodiester bond replaces by a covalent 5' phosphotyrosyl linkage. reaction haults at this point. additional proteins cooperate in the formation of an active Spo11 complex on DNA & in DNA processesing after it's cleaved |
|
|
processing of Spo11- mediated DSBs pt 1 |
1) complex of proteins bind to each spo11 complex & cleave DNA by several bp on 3' side liberating liked protein and segment of attached DNA strand |
|
|
processing of Spo11- mediated DSBs pt 2 |
2) nuclease Sae2 degrades DNA further, other enzymes are implicated to create long 3' ss extensions which can now be processed in a mechanism similar to DSB pair |
|
|
DSBR pathway in meiotic recombination |
1) ss regions are loaded onto 3' extensions on either side of DSB 2) site now set up for recombination 3) Stable DNA joint is then formed by intertwining of ssDNA w its compliment from the homologous target (recombinases involved in this step) 4) polymerase and ligase can complete the process of repair |
|
|
Catalysis of Holliday intermediate resolution |
RuvAB complex intermediate is resolved by nicking strand in each duplex followed by ligation processing of junctions is facillitated by RuvAB complex (repair of UV damage) up to 2 RuvA protein tetramers bind to form a complex w 2 RuvB hexamers |
|
|
RuvA |
protein holliday junction-specific DNA binding protein recognizes structure of DNA junctions and keeps it in 'box-like' state |
|
|
RuvB protein |
is a dna translocase Donut shaped hexamers surround 2 of the 4 arms of holliday intermediate DNA is propelled outward through the hole in the donut shaped RuvB away from the junction in a reaction coupled to ATP hydrolysis Results in very rapid movement of the position of the holliday intermediate (1000's bp in seconds) |
|
|
RuvC |
Holliday intermediate resolvase recruited after RuvAB complex moved junction away from damaged DNA RuvC replaces RuvA tetramer at junction & cleaves strands in opposing arms of intermediate by nicking 2 strands w same polarity, viable chromosomal product is formed RuvC does cleaving at site 1 or 2 |
|
|
Site-specific recombination |
precise & predictable DNA rearranged between 2 specific sequences involves movement of specialized nucleotide sequences called mobile genetic elements between non-homologous sites carried out by recombinase result in insertion, deletion or inversion |
|
|
Site-specific recombination effects |
alter gene order regulate gene expression & increase genetic repertoire in prokaryotes & eukaryotes Can result in spontaneous mutations |
|
|
Cre-LoxP & Flp-FRT transgenes |
LoxP & FRT are specific direction sequences places in genome (not naturally) Cre & Flp are recombinase enzymes that recognize LoxP & FRT sites respectively Cre & Flp recombinases not naturally occurring work to cleave or invert intervening sequence when engineering into the cells of any organism |
|
|
Outcomes of Cre-LoxP & Flp-FRT |
must be two LoxP or FRT sites in the genome for system to work depends on location & relative orientation of recombination sites in genomic DNA they reside |
|
|
Inverted Cre-loxP & Flp-FRT |
recombinase enzyme will invert the intervening sequence changes its orientation on the DNA |
|
|
Same direction orientation Cre-loxP & Flp-FRT |
recombinase cleaves out the intervening sequence leaving behnd reformed LoxP or FRT site |
|
|
Foreign DNA containing homolgy outside region of LoxP or FRT site
|
if this occurs & also contains 2 matching sites, intervening sequence of foreign DNA can be inserted to replace endogenous sequence between the sites |
|
|
The Brainbow technique |
makes use of green fluorescent protein (GFP) and site-specific recombination & traces path of neurons that make up the brain |
|
|
Zinc Finger Nucleases protein engineeirng |
protein domain characterized by a single atom of zinc coordinated to 4 cys residues or 2 his residues.
folds as alpha-beta-alpha residues within alpha helix structure are able to contact 3 consecutive nucleotides within major groove of DNA |
|
|
TALENs protein engineering |
DNA binding directed by series of TALE domains recognize single bp enzymes expressed in a cell & resulting enzyme cleaves the target site in the genome generating a ds break Designed to inactivate genes or integrate specific foreign DNA sequences |

