![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
18 Cards in this Set
- Front
- Back
|
What are the different types of bacterial movement? Give a brief description of each one. |
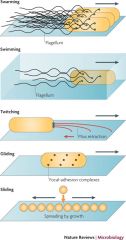
1. Swarming - multicellular movement across a surface 2. Swimming - individual movement of bacteria in liquid 3. Twitching - surface movemnt of bacteria powered by the extension of pili which subsequently retract 4. Gliding - active surface movement that does not require flagella or pili - rather with focal adhesion complexes 5. Sliding - passive surface translocation that is powered by growth and facilitated by secretion of surfactant |
|
|
What does 'swimming' movement require? |
Flagella |
|
|
What is the mechanism for 'twitching'? |
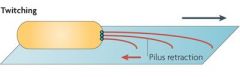
1. Pilus extension and polymerisation 2. Attachment to surface 3. Retraction of pili by depolymerisation |
|
|
What is the mechanism for 'gliding'? |
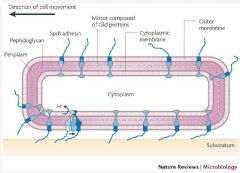
1. Large focal adhesion complexes extend from the cell and connect the extracellular surface to actin like cytoskeletal filaments. 2. Proteins attached to the intracellular portion of the FAC push backwards along the cytoskeletal filament and move the cell forwards whilst the FAC remain in position. |
|
|
Describe the structure of a flagella. |
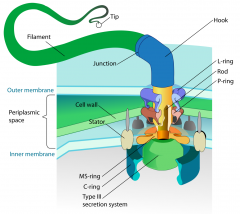
1. Basal body rooted in the cell membrane and cell wall 2. A hook anchored in the basal body 3. Long fine filament - a helical protein made up of flagellin subunits |
|
|
What causes the rotation of the basal body and the flagellum? |
Energy required to move a molecular motor through the proton motive force. H+ movement across the membrane through the 'mot' complex drives the rotation of the flagellum. |
|
|
What causes a bacteria to: 1. Run 2. Tumble ? |
1. CCW rotation = grouping of flagella to run 2. CW rotation = ungrouping of flagella to tumble |
|
|
Define chemotaxis. |
The ability of a cell to sense external concentration of a chemical species and migrate (directed) towards / away from higher concentrations |
|
|
Define phototaxis. |
Phototaxis = movement towards light - phototrophic organism orients itself most effectively for photosynthesis. |
|
|
Describe the movement of a bacteria when a chemical gradient does not exist. |
1. Runs - cell swims forward in smooth fashion 2. Tumbles - stops and jiggles about for a bit 3. Direction of next run is random 4. cell moves randomly but can go anywhere |
|
|
Describe the movement of a bacteria when a chemical gradient EXISTS. |
Random movements become biased. 1. Less tumbling longer runs 2. Organism moves up or down the concentration gradient towards / away from the chemical source |
|
|
How is the spinning direction of the flagella changed? |
Through the FliM switch protein. When the molecule 'CheY-P' binds to the FliM protein complex the protein switches motor direction to clock wise (tumbling) |
|
|
What are MCPs? |
Methyl acceptingchemotaxis protein which senses the presence or absence of attractants |
|
|
What is the role of CheW and CheA in the flagellar machinery? |
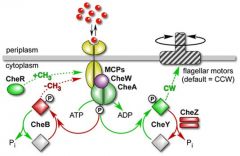
CheW with methyl accepting proteins regulate the activity of CheA - a histidine kinase. CheA autophosphorylates and goes on to activate CheY into its phosphorylated form CheY-P which induced tumbling. |
|
|
Describe in detail how the actions of the flagellar machinery changes in the presence of a chemical gradient. |
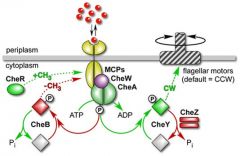
MCP's sense attractants which causes a conformational change which allows binding of CheW (thought to be a transducer) and CheA. CheA binding inactivates it resulting in no autophosphorylation and thus no phosphorylation of CheY. The lack of phosphorylation means that the FliM protein is not bound so biases running. |
|
|
Describe in depth the mechanisms which control MCP activity. |
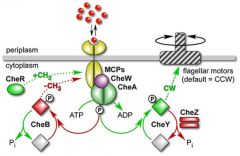
MCPs can be methylated by CheR - a methyltransferase MCPS can also be demethylated by CheB-P - a methylesterase. When MCPs are fully methylated they cannot respond to the presence of an attractant (outcome of CheR activity) When MCPS are demethylated by CheB-P they can sense and respond to the presence of the attractant. |
|
|
What phosphorylates CheB? |
CheA |
|
|
What causes running to cease? I.e why do bacteria run and then tumble momentarily when moving in response to a chemical gradient. |
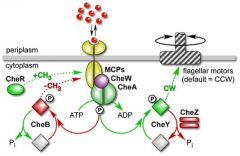
The two systems which regulate MCP wrestle when an attractant is sensed. 1. Initially the lack of kinase activity of CheA and therefore no phosphorylation/activation of CheB allows CheR to methylate MCPs - no attractant response 2. This means that the activity of CheA is off and therefore no phosphorylation of of CheB which allows CheR to methylate MCPs 3. Eventually this inactivates the MCPs, reactivating CheA which phosphorylates itself into CheA-P 4. CheA-P phosphorylates both CheY which causes tumbling and CheB which begins to demethylate the MCP again. 5. CheB reactivates the MCP, CheA activity is halted and running begins again. |

