![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
90 Cards in this Set
- Front
- Back
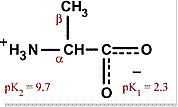
|
Alanine
Ala A Hydrophobic and nonpolar, mostly on interior of proteins Aliphatic |
|
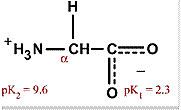
|
Glycine
Gly G Smallest amino acid, doesn't really have an effect on hydrophobic interactions Can be found anywhere on the protein |
|
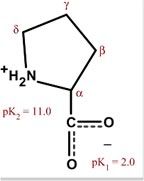
|
Proline
Pro P Non-polar Alipathic Constrains proteins, often found at end of alpha helices; or as a kink in alpha helix |
|
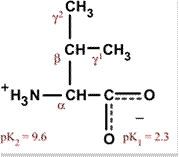
|
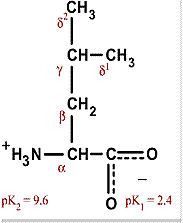
Valine
Val V Non-polar and hydrophobic, usually in interior Aliphatic |
|
|
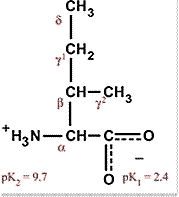
Leucine
Leu L Non-polar, hydrophobic, generally in center of protein Aliphatic Most common amino acid |
|
|
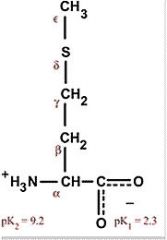
Isoleucine
Ile I Non-polar and hydrophobic, interior of proteins Aliphatic Usually interchangable with leucine or valine |
|
|
Methionine
Met M Non-polar and hydrophobic Always the first amino acid coded for by DNA to start a protein |
|
|
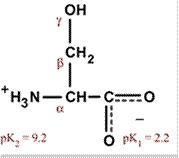
Serine
Ser S Polar, uncharged, hydrophilic Easily hydrogen bonds |
|
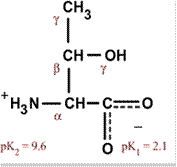
|
Threonine
Thr T Polar, uncharged, hydrophilic |
|
|
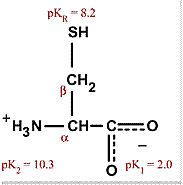
Cysteine
Cys C Polar, uncharged, hydrophilic Can form di-sulfide bonds unlike Met Means it is often found in secreted proteins since extracellular environment allows disulfide bonds |
|
|
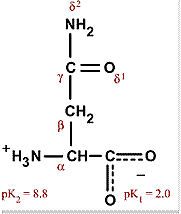
Asparagine
Asn N Polar, uncharged, hydrophilic Common site for attachment of carbohydrates in glycoproteins |
|
|
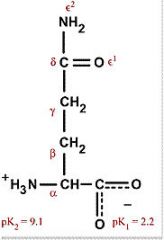
Glutamine
Gln Q (Qtamine) Polar, uncharged, hydrophilic |
|
|
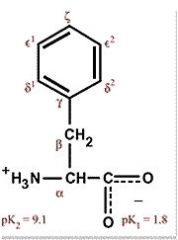
Phenylalanine
Phe F Nonpolar, aromatic, hydrophobic Absorbs ultraviolet light Often found in folded proteins since pi electrons stack with other aromatic systems |
|
|
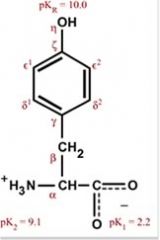
Tyrosine
Tyr Y Polar (weakly), aromatic, uncharged Absorbs ultraviolet radiation |
|
|
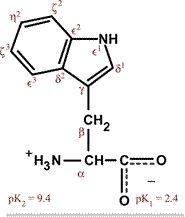
Tryptophan
Trp W (Twiptophan) Largest amino acid Polar (weakly), aromatic, uncharged Absorbs ultraviolet radiation |
|
|
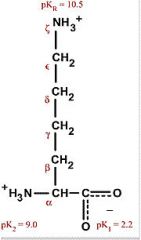
Lysine
Lys K (closest to L) Polar, positively charged, hydrophilic Still has some hydrophobic character to it |
|

|
Histidine
His H Polar, positively charged, hydrophilic Facilitate many enzyme-catalyzed reactions |
|
|
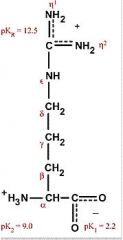
Arginine
Arg R Polar, positively charged, hydrophilic Found in nitrogen metabolism and phosphorylated substances |
|
|
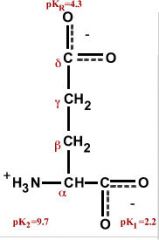
Glutamate (glutamic acid)
Glu E (Glutameke) Acidic, polar, negatively charged, hydrophilic |
|
|
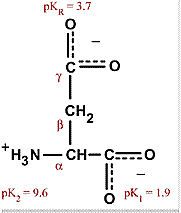
Aspartate
Asp D (Aspardic acid) Acidic, negatively charged, polar, hydrophilic Maintains solubility and ionic character of proteins |
|
|
1. Define amino acid residue.
2. Which amino acid is not chiral? |
1. When an amino acid is part of a protein chain, it is known as a residue since the peptide backbone is the main structure.
2. Glycine G (Gly) |
|
|
1. What is the D,L system? Draw them.
2. Which type are all amino acids? |
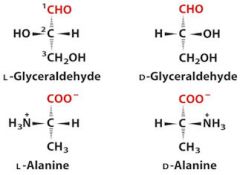
2. All amino acids are L.
|
|
|
Define absorbance.
|
The Lambert-Beer law defines it as e*c*l where c is the concentration and l is the path length. This means that absorbance is directly related to concentration.
Aromatic compounds often have a high absorbance. |
|
|
Define cystine.
|
When cysteine C (Cys) is oxidized to form a dimeric amino acid which are linked by a disulfide bond. These are strongly hydrophobic and nonpolar. They play a strong role in covalent bonds in tertiary structures.
|
|
|
Define zwitterion.
|
A substance that can act as either a base or an acid, has both a positive and negative charge at neutral pH. Like the peptide base of amino acids.
|
|
|
1. How does forming a zwitterion affect pKa?
2. Does a positively charged amino group give up a proton more easily or less easily when a negatively charged carboxyl group is present? |
1. This lowers the pKa because zwitterions are stable. Thus, the substance wants to move towards a zwitterion.
2. The amino group has a lower pKa because the oxygen pulls some electrons away from the amino group. |
|
|
1. What is the isoelectric point?
2. How do you calculate pI? |
1. Known as pI, it is the pH at which the net electric charge is 0.
2. Take the arithmetic mean of all the pKa values. For example, in glycine, you would combine the pKa of the carboxy group and the pKa of the amino group. |
|
|
1. If the pH > pI, what charge does the amino acid have?
2. If the pH < pI, what charge does the amino acid have? |
1. Negative
2. Positive |
|
|
1. How are peptide bonds formed?
2. Do peptide bonds last long? Why? |
1. By condensation reaction, removal of water from the carboxy group and amino group. Note that peptide bonds are not favored.
2. Peptide bonds have a high activations energy but a half life of 7 years. |
|
|
1. Define oligopeptide.
2. Define polypeptide. 3. Define proteins. |
1. Few amino acids
2. Molecular weight is less than 10,000 da 3. Molecular weight is greater than 10,000 da |
|
|
Describe the convention of displaying a polypeptide/protein.
|
Amino-terminal (N-terminal) is on the left, while carboxy-terminal (C-terminal) is on the right. The sequence is read left to right.
|
|
|
1. Give a range of molecular weights for proteins in our body.
2. Give a range of length for proteins in our body. |
1. 12,000 to 3 million
2. 100 to 27,000, but the majority are < 2,000. |
|
|
1. Define multisubunit protein.
2. Define oligomeric. 3. Define protomer. |
1. A protein with two or more polypeptides associated noncovalently.
2. If at least two of the polypeptides in a multisubunit protein are identical. 3. The identical polypeptides of a oligomeric protein. |
|
|
1. How do we estimate the number of amino acid residues in a protein given molecular weight?
2. How do we get this number? |
1. Divide molecular weight by 110 da.
2. Average molecular weight is 138. However, smaller amino acids predominate, so we drop this to 128. Then we have to take into account the condensation reaction, 110 da. |
|
|
1. Define conjugated proteins.
2. Define prosthetic group. 3. Give some examples of a conjugated protein. |
1. A protein that contains permanently associated chemical components that are not amino acids.
2. The non-amino acid part of a conjugated protein. 3. Lipoprotein, glycoprotein, metalloproteins |
|
|
What is the primary structure of a protein?
|
The sequence of amino acids; the description of all covalent bonds including disulfide and peptide bonds in an amino acid
|
|
|
What is the secondary structure of a protein?
|
Stable arrangements of amino acid residues giving rise to recurring structural patterns.
Spatial arrangement of backbone without regard to the side chain conformations. |
|
|
1. What is the tertiary structure of a protein?
2. What is the quaternary structure of a protein? |
1. Three dimensional structure of the entire polypeptide chain.
2. The spatial arrangement of two ore more subunits of polypeptides. |
|
|
1. Do proteins with different functions always have different amino acids sequences?
2. If you alter the primary structure of the amino acid, will the function of the protein change? |
1. Yes
2. Possibly. |
|
|
1. Is the amino acid sequence always the same for a particular protein?
2. Why are there diseases associated with these polymorphic proteins then? |
1. No, an estimated 20% to 30% of proteins are polymorphic, with different amino acid variations in the human genome.
2. There are critical areas of the protein, where the sequence must be conserved. |
|
|
1. Draw a peptide bond.
2. What conformation predominates? Trans or cis? |
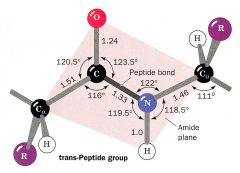
2. Trans. Cis can happen with proline in beta chains sometimes.
|
|
|
1. Define native protein.
2. Define stability as related to proteins. |
1. A protein in it's functional folded conformation.
2. The tendency for a protein to maintain a native conformation. |
|
|
1. What type of interactions predominate to fold proteins?
2. Of these, which predominates? |
1. Weak interactions like hydrogen bonding, hydrophobic and ionic interactions.
2. Hydrophobic |
|
|
1. What angle is Phi (circle with slash through it)?
2. What angle is Psi (trident)? 3. What angle is Omega (w)? |
1. C - N - Calpha - C
2. N - Calpha - C - N 3. Calpha - C - N - C (99% of the time trans) |
|
|
1. Define Ramachandran plot.
2. What is a regular secondary structure? |
1. The possible angles of Psi versus Phi for a protein configuration. Anything in blue is possible, yellow is not.
2. Has repeating angles. Two common types: alpha helix and beta sheets. |
|
|
Describe an alpha helix.
|
Polypeptide backbone tightly wound around an imaginary axis with the R groups pointing outwards.
|
|
|
1. Is an alpha helix right or left handed?
2. Why is an alpha helix so stable? |
1. Right handed, as your fingers curl, must trend upward. Right handed seems to be slightly more stable than left handed.
2. The amino acids on top of each other hydrogen bond. The hydrogen attached to the nitrogen group binds with the oxygen 4 amino acids away. |
|
|
1. How many amino acid residues are in every turn of an alpha helix?
2. How high is each turn of an alpha helix? |
1. 3.6 residues
2. 5.4 angstroms |
|
|
What are 5 constraints that affect the stability of an alpha helix?
|
1. Intrinsic propensity of a residue to form an alpha helix.
2. Interactions between R groups 3 to 4 residues apart. 3. Bulkiness of R groups, or electrostatic repulsion 4. Occurrence of proline or glycine 5. Interactions at the end of the helix which is a dipole |
|
|
1. Why does proline affect alpha helices?
2. Why does glycine affect alpha helices? |
1. Proline causes a kink in the alpha helix with the rigid nitrogen bond. Also, there is no hydrogen on the nitrogen to participate in normal hydrogen bonds of the alpha helix.
2. Glycine is so tiny that it has more flexibility and forms a different coiled structure. |
|
|
Describe a beta strand.
|
Zigzag polypeptide chains that resemble sheets. Hydrogen bonding happens between the hydrogen attached to the nitrogen and the carbonyl group. The R groups alternate pointing up and pointing down.
|
|
|
Describe the two types of beta strands.
|
Antiparallel is where the polypeptides run antiparallel to each other.
Parallel is where the polypeptides run parallel to each other. |
|
|
Describe a beta turn.
|
A four amino acid sequence connection between anti-parallel strands of a beta pleated sheet. Allows a 180 degree turn. Typically involve either proline P (pro) or glycine G (gly) since P participates in cis bonds and glycine is super small. It is a non-repetitive structure.
|
|
|
Describe an omega loop.
|
It is a non-repetitive structure that is compact and globular. R groups are inside.
|
|
|
What is circular dichroism (CD) spectroscopy?
|
Plots absorption spectrums for proteins. Thus, we can determine if the proteins are properly folded, estimate the percentage of the protein that is folded in the common secondary structures and monitor transitions between folded and unfolded states.
|
|
|
1. Define fibrous protein.
2. Define globular proteins. |
1. Polypeptide chains arranged in long strands or sheets. Generally a single secondary structure and a simple tertiary structure.
2. Polypeptide chains folded into a spherical or globular shape. Several types of secondary structures usually. Typically enzymes or regulatory proteins. |
|
|
Describe alpha-keratin.
|
Found only in mammals, this is a type of intermediate filament. Two right handed alpha helices are wound together in a left handed alpha helix. The interior of this helix is full of hydrophobic R groups. These interactions and disulfide bonds help stabilize the structure.
|
|
|
1. Define dimer as it relates to alpha-keratin.
2. Define protofilament. 3. Define protofibril. 4. Define microfibril. |
1. Two polypeptide strands wound around each other in a left-handed helix.
2. 3 of these dimers strung together. 3. 12 of these dimers strung together, or 4 protofilaments packed together. 4. 8 of these protofibrils strung packed together. |
|
|
How does a perm work?
|
Reduce the disulfide bonds, curl the hair and reform the disulfide bonds. Now, when original alpha helix formations form, these new disulfide bonds will torque the hair.
|
|
|
Describe collagen.
|
A strong tensile fiber found in tendons or bone matrix. Consists of 3 alpha chains (not helices) which are left handed, supertwisted together right-handedly. Has 3 amino acids residues per turn and the sequence is usually glycine, proline then 4-Hyp (an uncommon amino acid).
|
|
|
Why does having Vitamin C protect you against scurvy?
|
Scurvy is the weakening of collagen strength, which eventually causes death. Both Pro and 4-Hydroyproline are needed in collagen. Alpha-ketoglutarate is consumed converting Pro -> 4-Hyp, but it is also consumed in another reaction with iron. Vitamin C is needed to reduce the heme iron so that the enzyme can work again.
|
|
|
What type of covalent crosslinking does collagen have?
|
Not disulfide bonds, but lysyl oxidase bonds which use Lys and HyLys (5-hydroxylysine)
|
|
|
Describe the structure of silk.
|
Silk is made of fibroin protein. Almost all of it is in the beta conformation (anti-parallel). Silk does not stretch, but will bend since there are extensive hydrogen bonds between the sheets.
|
|
|
1. What are the effects of non-polar R groups in tertiary structure?
2. Charged polar R groups? 3. Uncharged polar R groups? |
1. Interior, hydrophobic interactions.
2. Exterior, ionic interactions. 3. Either, hydrogen bonding, |
|
|
Describe myoglobin.
|
Small globular protein. 153 amino acids, 16.7 kDa and has 8 alpha helices. Hydrophobic proteins packed in core (only room for 4 water molecules), peptide bonds in trans, and proline present at many of the turns and 1 at strategic kink in an alpha helix. 3/4 residues per turn.
|
|
|
Define motif.
|
Also known as supersecondary structure or fold, it is a recognized folding pattern involving two or more elements of secondary structure and connections between. Has functional significance. Examples are B-a-B loop or a-a corner.
|
|
|
Define x-ray diffraction.
|
Place a crystal of protein (limitation of crystal) between x-ray and source and measure the reflections using mathematical Fourier transforms. Can be applied to proteins too large for NMR.
|
|
|
Define NMR.
|
Works on proteins dynamically. When a magnetic field is applied, some atoms give rise to a magnetic dipole which shows up on a graph. Using computers, we can then use this info along with van der Waals radii, etc to plot the 3-D structure of the protein.
|
|
|
Define domain.
|
Describes structural patterns in a protein. Part of a polypeptide chain that is independently stable or could undergo movement as a single entity with respect to the entire protein. Different domains often have different functions.
|
|
|
Define Rossman fold.
|
1. A type of motif that has dinucleotide binding (for DNA).
2. Includes a B-a-B loop that then has a bunch of beta segments attached to it's neighbor by an alpha helix segment. Found in enzymes, often with a binding site. |
|
|
List 5 rules for protein folding.
|
1. Hydrophobic R groups bured in core.
2. B and a found in different layers. 3. Segments close to each other in sequence are usually close together in folded. 4. Connections of elements do not typically for knots or crossover 5. B most stable with slight right twist. |
|
|
1. What are the categories in the Structural Classification of Proteins?
2. Define protein family. |
1. All a, all B, a/B (integrated) and a + B (alpha and beta somewhat seperated).
2. Proteins similar in primary structure of tertiary structure and function. |
|
|
Define superfamilies.
|
Protein families with similar function that use the same motif. Probably evolutionarily related.
|
|
|
Why did domains evolve?
|
They are stable folding patterns, which can tolerate amino acid changes. They support function, thus often domains are conserved through evolution, not specific primary structure.
|
|
|
1. Define multimer.
2. Define oligomer. 3. Define protomer. |
1. Multisubunit protein.
2. Multimer with just a few subunits. 3. Repeating structural units in a multimeric protein. |
|
|
Go into more detail about quaternary structure and symmetries.
|
Subunits are symmetrically arranged and associate non-covalently.
Rotational or helical symmetry means that subunits can be superimposed on others by rotation around some axis. |
|
|
List four forces that stabilize protein structure.
|
1. Electrostatic forces
2. Hydrogen bonding forces 3. Hydrophobic forces 4. Chemical crosslinks |
|
|
1. What are salt bridges?
2. Define hydropathies. |
1. Electrostatic interactions between two ionic groups of opposite charges within a protein.
2. Combined hydrophilic and hydrophobic tendencies of individual residues. High value means more likely interior. |
|
|
1. Define denaturation.
2. What are some things that cause denaturation? |
1. Loss of 3D structure enough to affect function. Cooperative process, has a sigmoidal curve, denaturation happens all at once.
2. Extreme heat, pH or salt concentrations. |
|
|
Define renaturation.
|
If a denatured protein is returned to conditions that are favorable for function, sometimes the protein will return to the stable native conformation.
Ribonuclease A can be renatured. Has 4 disulfide bonds that can be broken and remade. |
|
|
Describe the two hypotheses of protein folding.
|
1. Folding process is hierarchical. First comes a and B structures, then ionic interactions help things, etc.
2. Folding is initiated by a spontaneous collapse of the polypeptide into a compact state (molten globule) |
|
|
Describe a free energy funnel.
|
Free energy decreases as you move down the graph, shows how a protein folding would act. Decreases entropy and increases proteins in native conformation as move down. There is also a reduction of possible conformations.
|
|
|
Define molecular chaperone.
|
Proteins that interact with partially folded or improperly folded proteins. Prevent aggregation, assist in assembly, and allow refolding. Examples include Hsp70, GroEL/GroES and enzymes.
|
|
|
Define Hsp70.
|
A molecular chaperone abundant in cells stressed by elevated temperatures. Bind to hydrophobic regions to prevent aggregation, block folding until translocated through membrane, and assist in general assembly. ATP dependent.
|
|
|
Define GroEL and GroES chaperones.
|
Fold proteins that do not fold spontaneously. Up to 10% of proteins in E.coli need this. They work together and are ATP dependent.
|
|
|
Define PDI.
|
Protein disulfide isomerase is an enzymatic molecular chaperone that rearranges disulfide bonds.
|
|
|
Define PPI.
|
Peptide prolyl cistrans isomerase is an enzymatic molecular chaperone that converts between cis and trans isomers of proline.
|
|
|
1. Define amyloid.
2. Define amyloidoses. |
1. A protein that has been secreted in a misfolded state and has been made into an insoluble extracellular fiber.
2. Diseases associated with amyloids. |
|
|
1. Define primary amyloidosis.
2. Define secondary amyloidosis. |
1. Independent of any other disease, like immunoglobin light chain accumulation.
2. Accompanied by a long term illness like RA, tuberculosis, cancer. |
|
|
1. How can amyloidosis affect diabetes?
2. Alzheimer's? 3. Parkinson's? |
1. Amylin forms on pancreatic islets cells, eventually destroying them.
2. Amyloid plagues, presenillin build up in extracellular spaces while tau fibers build in intracellular neuron spaces. 3. Intracellular fibers called Lewy bodies build up. |

