![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
108 Cards in this Set
- Front
- Back
- 3rd side (hint)
|
Metabolic pathways hpic |
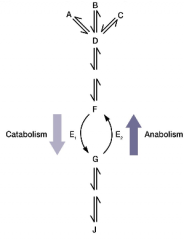
Metabolism - all chemical reactions in a cell Catabolism - breakdown of complex molecules into smaller ones with the release of energy for Anabolism - the reactions that build microbial cells |
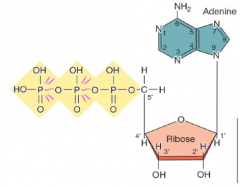
ATP is the energy currency of cells |
|
|
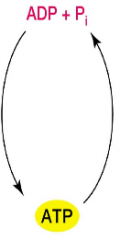
Cells energy cycle (hpic) |
Aerobic respiration: Oxygen is used to make ATP Anaerobic respiration: other than oxygen is used Fermentation: substrate level phosphorylation, how fermentation makes ATP Photosynthesis, using light energy to make ATP |
|
|
|
Energy provided in a cell |
we can predict the direction of a reaction using a quantity - Gibbs free energy change ∆G - ∆G for a reaction determines how much energy is available to do work such as: –Rotate a flagellum –Build a cell wall, or –Store information in DNA |
|
|
|
Reaction mechanism and Keq |
![Keq= [C][D]/[A][B]
∆G=-2.303RT log(Keq)
∆G units= Kcal/mol or Kjoules/mol](https://images.cram.com/images/upload-flashcards/70/43/24/6704324_m.png)
Keq= [C][D]/[A][B] ∆G=-2.303RT log(Keq) ∆G units= Kcal/mol or Kjoules/mol |
|
|
|
Unbalanced reaction mech |
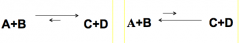
Left: Keq>1 ∆G = is negatve, E is released Right: Keq<1 ∆G = is positive, E was required (endergonic) |
|
|
|
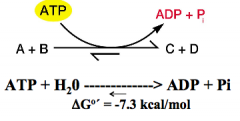
ATP input makes endergonic reactions more favorable |
|
|
|
|
Oxidation-Reduction (Redox) Reactions and Electron Carriers |
-electrons move from donor to acceptor -utilize carriers (to move electrons) -redox reactions can result in energy release, which can be used to form ATP |
|
|
|
Oxidation-Reduction Reactions (hpic) |
-oxidation = loses electrons -reduction = gain electrons -oxidized substance is the donor, and the reduced substance was the acceptor (redox couple) *easy stuff, you better not forget |

Krebs cycle (TCA cycle) malate gets oxidized, NAD is reduced, NADH is the cell reducing power |
|
|
Rhodoferax (hnotes) |
Rhodoferax metabolins is psychrophillic, strictly anaerobic bacterium that oxidizes Acetate with the reduction of Iron (psychrophillic, survives in cold temps) oxygen is toxic to it. survives where Atmosphere doesnt reach |
-uses Acetate as electron donor (its being oxidized) dumps electrons onto Iron. -Malate dehydrogenase: the enzyme that oxidizes the Malate |
|
|
Enzymes (hpic) |
-proteins (mostly) that catalyze reactions( ribozymes = catalytic RNAs) -Activation energy: the energy required to bring reacting molecues together, (enzymes lower this Act-Energy) -Often named after the reactions they catalyze ex( phosphatase, kinase, cellulase) |
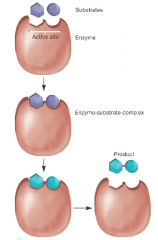
-increase local concentrations of substrates -orientation substrates for proper reaction |
|
|
Induced fit model of enzymes (hpic) |
1) ATP and glucose bind to active site of enzyme 2) enzyme undergoes conformational change that makes substrates fit the mold 3) substrates are converted into products 4) release products |
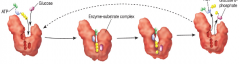
|
|
|
Reduction potential: (Eg) the ∆G equivalent (hnotes) |
- Equilibrium constant for redox rxns (Eg is like ∆G) -measure tendency of donor to lose electrons more negative Eg = better donor -more positive Eg = better acceptor |
redox couples with more negative Eo, will donate electrons to more positive Eo |
|
|
The Electron tower: plot/chart of Eo values |
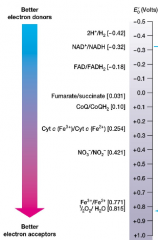
|
|
|
|
2 classes of electron carriers in redox reactions (hnotes) |
-freely diffusible: ex_ NAD+ and NADP+ -membrane-bound: ex) flavoproteins, cytochromes, quinones |
carriers function togheter moving electrons from donors to acceptors. AKA electron transport systems (most are found and formed in the plasma membrane, not the cytoplasm or cell wall) |
|
|
Types of diffusible electron carriers |
NAD+ and NADP+ (NADH and NADPH are the reducing power of the cell)
|
NAD+ = nicotinamide adenine dinucleotide NADP+ = nicotinamide adenine dinucleotide phosphate |
|
|
Types of membrane bound electron carriers |
Quinones: ex) Coenzyme Q Cytochromes: are proteins, use iron to transfer electrons (iron is part of a heme group)
|
|
|
|
Mircrobes transfer energy by moving electrons from: |
reduced food molecules --> carriers in cytoplasm (NADH) --> membrane-bound carriers, (ex cytochromes, quinones) latch onto O2, metals, and oxidized Nitrogen and sulfur Summary: gain energy from movement of electrons through redox rxns |
|
|
|
2 metabolic groups of the carbon cycle |
Heterotrophs -reduced, preformed organic compounds as Carbon source -convert large amounts of C --> CO2 (ex humans, many microbes) Autotrophs: use CO2 as carbon source -ex: plants and microbes -produce organic compounds used by heterotrophs eg. sugar |
|
|
|
Organisms that require sources of energy and electrons for growth |
Phototrophs: use light for energy
Chemotropths, ozidize chemical compounds, often same as their carbon sources |
|
|
|
Microorganisms have 2 sources for electrons |
lithotrophs: inorganic molecules as electron donors organotrophs: organic molecules as donors for electrons ultimate electron acceptors: -inorganic molecules in Respiration -Organic molecules used in Fermentation |
|
|
|
Quiz: you identify a bacterium that utilizes compounds as a source of carbon, energy, and electrons, what is this bacterium? |
ANS: chemo-organo-heterotroph.
|
Table 11.2 in book. test yourself |
|
|
nutritional types of organisms |
Almost all Eukarya are either photoautotrophs (plants/algae) or heterotrophs( anmals, protozoa and fungi) **Lithotrophy is unique to a few bacteria and archaea
|
|
|
|
Fueling reactions |
Despite diversity of energy, electron and carbon sources used by organisms, all have same basic needs: -ATP is energy supply -reducing power to supply electons for rxns. -precurser to biosynthesis |
|
|
|
Energy source (organotrophs) hpic |
ATP by 2 means: Substrate level phosphorylation
oxidative phosphorylation :ADP --> AGP |
|
|
|
two functions of organic energy sources hpic |
Amphibolic pathways: a pathway that works catabolic and anabolic. oxidized to release energy these are building blocks for anabolism. |
|
|
|
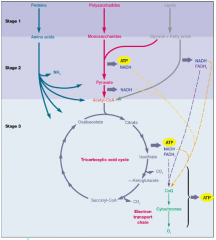
Aerobic respiration hnotes |
process that can completely catabolize an organic energy source to CO2 using: -glycolytic pathways (glycolysis) -tricarboxylic acid cycle -electron transport chain with oxygen as final electron acceptor. -produces ATP (indirectly by E-transport) |
normally uses glucose as carbon source
|
|
|
breakdown of glucose to pyruvate (3 pathways) know key rxns and products of each pathway |
embden-meyerhof (glycolysis)
pentose phosphate pathway
Entner-doudoroff pathway |
|
|
|
embden-meyerhof (glycolysis) hpic |
most common form of glucose breakdown occurs in cytoplasm functiosn in presence or absence of O2 has 10 rxns in two stages (6 carbon stage and 3 carbon stage) |
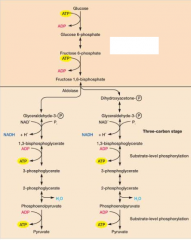
6C stage: glucose is phosphorylated twice, generating fructose 1,6-biphosphate 3C state: fructose 1,6 bisphosphate, spits into 3 glyceraldehyde then converted into pyruvate |
|
|
3-C stage indepths hpic |
key steps- oxidations come from NADH as reducing power substate level phosphorylations generate ATP net yield: 2 ATP, 2 NADH, and 2 pyruvate |
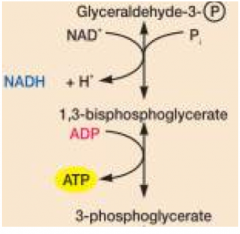
G3p oxidized and phosphorylated generates high energy p-bond |
|
|
pentose phosphate: prokaryotes and eukaryotes do this |
starts by converting Glucose-6-phosphate into ribulose-5-P generates sugars for biosynthesis (transketolases, transaldolases) -1ATP and 6NADPH is the reducing power for biosynthesis |
|
|
|
Entner-doudoroff (found in a few prokaryotes, not in euqkaroytes) |
combines rxns of glycolysis and pentose phosphate generates 1ATP, 1NADH, 1NADPH
|
|
|
|
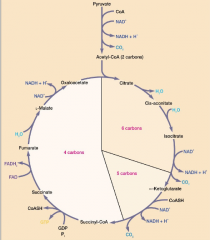
Tricarboxylic acd (krebs) cycle |
pyruvate is completely oxidized to CO2 happens in mitochondria of eukaryotes, and the cytoplasm of prokaryotes. Generates: CO2, NADH, FADH2 |
|
|
|
TCA cycle hpic |
-pyruvate first oxidizes CO2 and acetyl CoA, -acetyl-CoA condensed with oxaloacetate -oxidation and decarboxylation forming NADH and CO2 |
|
|
|
Electron transport and oxidative phosphorylation |
only 4 ATP molecules synthesized directly from oxidation of glucose most ATP is made when NADH and FADH oxidzed in electron transport chain |
|
|
|
Electron tranport chain hpic |
electrons from NADH and FADG are terminal acceptors they will flow from carriers with more negative energy to more positive energy and the energy released is used to make ATP by Oxidative phosphorylation -3ATP per NADH using O2 |
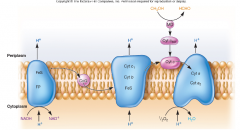
|
|
|
Oxidative phosphorylation |
Chemiosmotic hypothesis: - energy released during e-transport is used to establish proton gradient and charge difference across membrane -AKA Proton Motive Force (PMF) **memorize!! |
PMF: drives ATP synthesis e-flow causes protons to move outward across membrane, ATP is made when they move back in ATP synthase (enzyme) uses proton movement to catalyze ATP synthesis |
|
|
ATP synthese (F0, F1) hpic |
we use proton flow down the gradient to make ATP top half (F0) contains a proton channel, ring of C-subunits rotates Bottom half (F1) sphere (gamma) shaft will rotate, causes conformational changes in alpha and beta subunits, this synthesizes ATP from ADP |
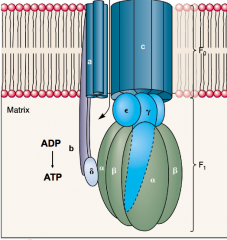
|
|
|
"Shewanella" electron transfer (an example of a microbial fuel cell) hpic |
this bacterium is aquatic, gram negative, and can transfer electrons to extracellular metals. must be in a Anoxic environment (low O2, the anode) it will connect to a chamber that is Oxic (high O2, the cathode) and a wire will connect the two chambers, circuit is generated. This is known as a microbial fuel cell |
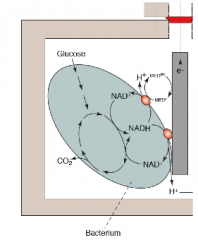
|
|
|
methods and types of Electron acceptors microbes use (hpic) |
Organic electron donor: -fermentation: endogenous electron acceptor (ex: pyruvate) -Aerobic respiration: O2 -anaerobic respiration: NO3, SO4, CO4, fumerate Inorganic E donor: chemolithotrophy: O2, SO4, NO3 (exogenous electron acceptors, ex) O2, NO3, SO4, CO4, Fumerate)) |
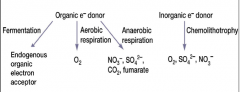
|
|
|
Anaerobic Respiration |
Less ATP is generated anaerobically (influenced by reduction potential) "Paracoccus": uses NO3 as terminal e- acceptor "geobacter" uses Fe3+ in anaerobic environments Denitrification: dissimilatory nitrate reduction: nitrate as terminal e- acceptor, becomes reduced to N2 gas) causes loss of Nitrogen in soil |
|
|
|
Fermentation "the consequence of life without air" hpic |
completion of catabolism without the electron transport system and a terminal electron acceptor. -Occurs in the cytoplasm -Hydrogen from NADH transfers onto pyruvate -generates: fermentation products(lactic acid, ethanol)also NAD+, -ATP by SLP (substrate level phosphorylation)
|
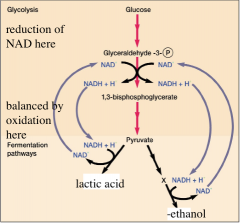
|
|
|
Chemolithotrophs hnotes |
acquire e-from the oxidation of inorganic sources such as H2, NO2 or Fe2+ the e- are transferred to terminal (exogenous) acceptors (O2) by e- transport chains |
2 examples: iron oxidizing and nitrifying |
|
|
Iron-oxidizing Bacteria ex) acidithiobacillus ferroxidans |
-oxidizes ferrous Fe2+ to ferric Fe3+ -O2 as an e- acceptor -forms insoluble ferric hydroxide
|
|
|
|
Nitrifying Bacterium |
obligate aerobes nitrification: oxidation of ammonia into nitrate -2 genera (nitrosomonas - ammonia to nitrite) (nitrobacter - nitrite --> nitrate) this is used to remove ammonia in waste water denitrification: nitrate back into N2 gas |
|
|
|
Sulfolobus (domain archaea) |

H2S---> H2SO4 |
|
|
|
Photosynthesis - phototrophs hnotes |
Two parts Light reactions: light every trapped, and converted into chemical Dark reactions :(calvin cycle) used to reduce CO2 and synthesize cell material (many phototrophs are also autotrophs) |
oxygenic: oxidize H20 for electrons, form oxygen. this is also called oxygenic photosynthesis. done by eukaryotes and cyanobacteria anoxygenic photosynthesis: electrons from other sources, done by all other types of bacteria |
|
|
Light reactions (trapping light) hpic |
Chlorophylls: Major light abosrbing pigments. eukaryotes, and cyanobacteria do this bacteriochlorophylls: major light absorbing pigments, done by purple and green bacteria |
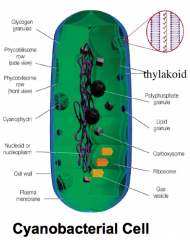
gram negative: because outer membrane(lipid bilayer) |
|
|
Domain "bacteria" Genus: prochlorococcus hpic |
Habitat: tropical oceans the rings in the picture are the thylakoids (where photosynthesis takes place) >100,000 cells/ml smallest known photosynthetic organism small genome 2000 parts) |
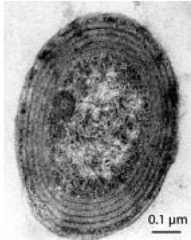
|
|
|
Accessory pigments hpic |
-transfers light to chlorophylls -absorb different wavelengths than chlorophylls -quench toxic forms of oxygen (photoprotection, antioxidants) increase the number of wavelengths that can be absorbed -ex) carotenoids, phycobiliproteins |
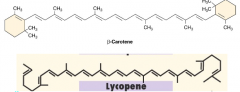
|
|
|
Photosystems |
when chlorophylls and accessory pigments are assembled into light harvesting arrays. these are imbeded in the thylakoids. 2 types, photosystem 1 and photosystem 2 |
|
|
|
Light reactions: green plants and cyanobacteria (hpic)
|
photosystem 1 absorbs P700 (cyclic phosphorylation) photosystem 2 absorbs P680 (non-cyclic phosphorylation) electron from PS1 travels down a electron transport chain to PS2, then back to the beginning of PS1. the electron can also travel in a different direction to make NADP+ into NADPH (calvin cycle) PS2 absorbs 2 protons, goes up in redox potential, and feeds electrons into PS1. |
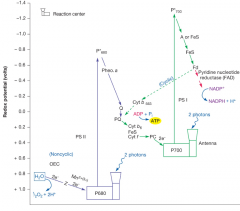
|
|
|
Photosystem 1 & 2 diagrams |
Use the inward flow of protons to synthesize ATP. |
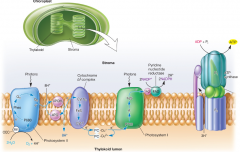
|
|
|
Light reactions: green and purple hpic |
The reactions occur in the plasma membrane -utilizes bacteriochlorophyll (not chlorophyll) to generate energy -Anoxygen (doesnt use water as reactant) -only one photosystem (restricted to only cyclic phosphorylation to make ATP) -Reverse electron transport (pushes electron backwards) to make NADP into NADPH -H2S or other organic donors are needed to replace the electrons |
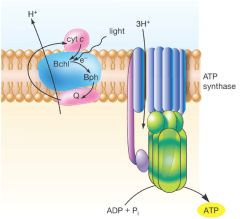
|
|
|
Archaea and photosynthesis? hpic of rhodopsim |
Some are photosynthetic, but lack both chlorophyll and bacteriochlorophyll. instead they use a protein pigment called Rhodopsin -there are 7 transmembrane helices. -conformational changes in rhodopsin- this is a proton pump, makes a proton motive force, generates ATP (normally this proton motive force comes from electron transport chain) |
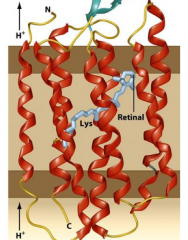
|
|
|
Metabolic reactions hpic |
metabolism –total of all chemical reactions catabolism –breakdown of larger,molecules into simpler ones –energy released used for work anabolism –synthesis of complex molecules from simpler ones with the input of energy |
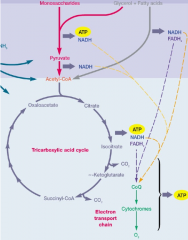
|
|
|
Anabolism hpic |
synthesis of complex molecules from simpler ones with the input of energy |
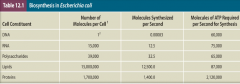
|
|
|
Principles of Anabolism hpic |
-Large from small -Many enzymes do double duty(catabolic), others function in only one direction (anabolic) -Catabolic and anabolic pathways use different cofactors -Catabolism produces NADH, NADPH -Anabolism uses NADPH as e donor |
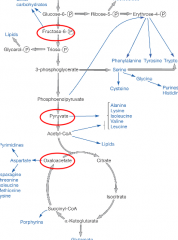
|
|
|
Calvin Cycle - Anabolic Pathway for Fixing CO2 Into Carbohydrate (hpic) AKA the dark reaction of photosynthesis |
AKA the dark reaction -energy demanding -occurs in chloroplasts in plants, and cytoplasm in bacteria -crucial for life, provides the organic matter for heterotrophs. -three phases: carboxylation, reduction, regeneration) |
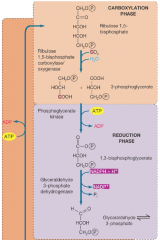
|
|
|
Calvin cycle energy path hpic |
-Biosynthesis requires energy and raw material -Materials are intermediates of central metabolic pathways -precursor metabolites (circled red) |
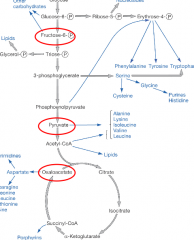
|
|
|
Reversal of catabolic pathways hnotes |
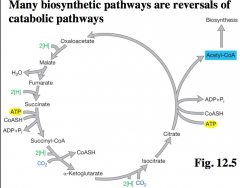
|
arrows are in reverse, instead of generating CO2, and power driven NADPH, its fixing CO2 to be used up. |
|
|
Carboxylation phase of calvin cycle hpic |
makes 3 phosphoglycerates this phase occurs in carboxysomes |
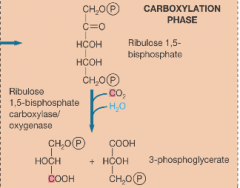
|
|
|
Reduction phase or calvin cycle hpic |
its the reverse of two key reactions in glycolysis. this step requires ATP |
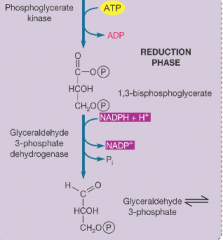
|
|
|
Regeneration phase of calvin cycle hpic |
Numerous carbohydrates are produced in this phase |
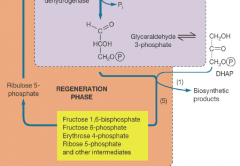
|
|
|
Gluconeogenesis hpic |
glucose from noncarbohydrate substrates -animals, plants, fungi, microbes use gluconeogenesis to maintain blood glucose levels -requires ATP and GTP -there are 6 enzymes also used in glycolysis, but also 4 that are unique to gluconeogenesis
|
|
|
|
Gene Structure and Replication hpic |
-DNA and RNA structure and roles -3 processes of genetic information flow: 1. DNA replication 2. Transcription 3. Translation |
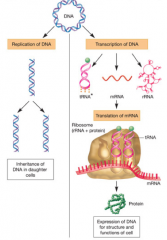
|
|
|
DNA as Genetic Material: Microbes (and some luck) Provided Proof, Griffith’s Transformation Experiments hnotes |
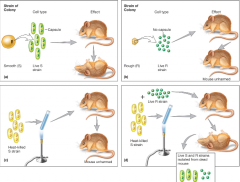
|
bacterium used in experiment streptococcus pneumoniae |
|
|
Terms for genomic vocab |
Gene – functional unit of genetic information – deoxyribonucleic acid (DNA) –Genes: pilA, lacZ – Proteins: PilA , LacZ •Genome – all genetic material in cell or virus – bacterial genomes consist of one (usually) or more DNA chromosomes genotype: specific set of genes carried in genome phenotype: observable characteristics |
|
|
|
Organization of cystronic RNA hpicRibonucleic acid (RNA) Structure |
promotor: where RNA polymerase binds to begin transcription operator: where repressor proteins bind to block transcription |
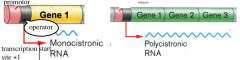
|
|
|
DNA Structure hpic |
polymer of nucleotides •each nucleotide 3 parts: –sugar-deoxyribose – nitrogenous base •adenine (A), guanine (G)-purines •cytosine (C), thymine (T) -pyrimidines •phosphate group •base + sugar + P group = deoxynucleotide |
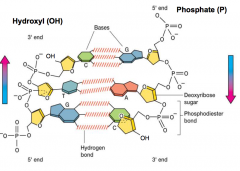
|
|
|
DNA structure part 2 |
Double helix, 2 complementary strands -each helix connected by phosphodiester bonds -sequence of one strand determines the other base pair rules ( A -->T 2H's & G --> C 3H's) |
|
|
|
DNA Size |
bacterial genomes vary greatly: –one of smallest - Mycoplasma - 580 kb encoding 480 proteins (lack a cell wall) expressed as base pairs (bp) 1000 bp=1kb -E. coli = 4,640kb or 4.64 Mb |
|
|
|
Ribonucleic acid (RNA) Structure |
3 types" - tRNA: carries amino acids during protein synthesis -rRNA: component of ribosomes (ribosomes + RNA) -mRNA: template for protein synthesis RNA: uses ribose vs deoxyribose, uracil replaces the thymine, usually singlestranded |
|
|
|
How is DNA Organized in Cells? |
Double helix in both prokaryotic and eukaryotic cells • However, organization differs |
|
|
|
Prokaryotic DNA hpic |
usually closed circular, supercoiled molecule -bacteria pack their DNA into loops or domains, collectively called the nucleoid |
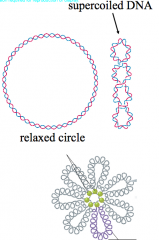
|
|
|
Eukaryotic DNA hpic |
-Linear -Eukaryotes wrap their DNA around proteins called histones, collectively called nucleosomes. -genes in human genomes are interrupted by introns ( archaea have circular chromosomes with histones) |
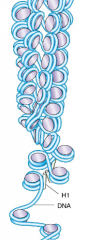
|
|
|
DNA replication hpic |
Semiconservative: -parental strands are conserved strands separate as templates for synthesis of new strands |
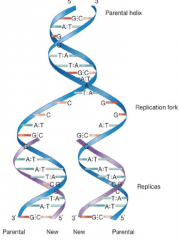
|
|
|
DNA replication in eukaryotes hpic |
-bidirectional - multiple origins of replication |
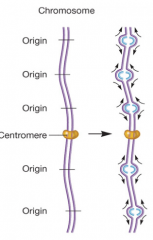
|
|
|
DNA replication in Prokaryotes hpic |
-bidirectional -single ori -2 forks moves in opposite directions How is ori selected? (DnaA is the DNA binding protein, initiates the opening at the site, begins the replication) |
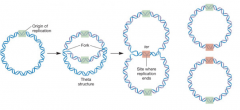
|
|
|
DNA polymerase |
Catalyzes DNA synthesis in 5' -->3' direction Needs these to function: - a template - deonxtnucleotide triphosphates (dNTP's) - Primer with 3' OH group (an RNA that is complementary to the DNA) - In bacteria, DNA Pol III is major DNA replication enzyme |
|
|
|
DNA polymerase pictures for both eukaryotes and bacteria hpic (SSB = single stranded binding proteins) part 1, then part 2 |
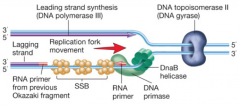
|
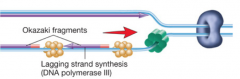
|
|
|
DNA gyrase (DNA topoisomerase II) hnotes |
DNA gyrase is the underwinder that is ahead of the replication fork. helps faciliate unbinding. the target for quinolone antibiotics |
DnaB Helicase: hydrogen bond breakers, is the thing that actually unwinds the DNA strand DNA Primase: it lays down RNA primer. |
|
|
Transcription of Bacteria |
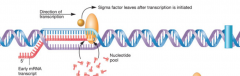
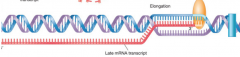
-RNA comes from DNA -RNA polymerase composed of = core + sigma factors -Sigma factors = proteins, that direct the core to the promotor, dictates where the RNA polymerase will bond - antibiotic "rifampin" targets bacterial RNA polymerase, disabling it -Generates 3 types of RNA: mRNA, tRNA, etc |
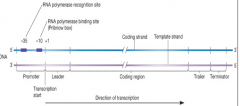
template strands direct rNA synthesis terminator is a DNA sequence that signals RNA pol to stop transcription |
|
|
What is a terminator? corresponds to picture in the next slide |
-it is a DNA sequence -broken into two groups: one group encodes an RNA stem loop structure. causes RNA pol to release (come off the template) protein Rho binds RNA, moves towards RNA pol and causes release |
|
|
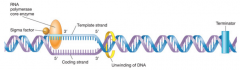
RNA polymerase "3 steps" (hpic) transcription |
|
|
|
|
Bacteria use two-component signal transduction systems to control gene transcription in response to their environments |
Signal, enters sensor kinase, requires ATP to power the kinase domain. histodine will grab the phosphate group that got bumped off. the phosphate will phosphorylate Aspertate, and activate a protein(response regulator) will interact with gene. |
|
|
|
Transcription - Eukarya hpic |
takes place in the nucleus -has 3 RNA polymerases -dont have sigma factors, but instead transcription factors -TATA box, promoter element. -RNA splicing to remove introns -mRNA further modified, also has capping, Methylguanosine added at the 5' end, polyadenylation: adenine nucleotides add (cap) to the 3' tail end |
|
|
|
Translation |
-synthesis of polypeptide directed by nRNA sequence - requires ribosomes and energy in the form of ATP and GTP (guanosine triphosphate) bacterial ribosomes: 2 subunits, 30S + 50S. the 30S = 21 proteins + 16 rRNA the 50S= 34 proteins + 23S and 5S rRNA |
|
|
|
rRNA's play important roles in translation hnotes |
-23S rRNA: peptidyltransferase, Ribozyme( participates in translation), -16S rRNA: helps aligns mRNA with ribosomes so translation can proceed, has sequence complementary to Shine-Dalgarno sequence of the mRNA |
|
|
|
ribosomes: sites of protein synthesis
|
-ribosomes "read" mRNA sequence as a code Codons: -3 nucleotides (triplet) -there are 64 codons - 61 specify amino acids (sense) -3 are stop codons (nonsense) -code is degenerate- multiple codons can encode the same amino acid. |
|
|
|
tRNA (transfer RNA) molecules hpic |
Convert and transfer aminoacids to ribosomes: -convert of the language of RNA into that of proteins -clover leaf shape -two functional regions |
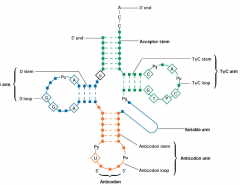
|
|
|
translation initiation hpic |
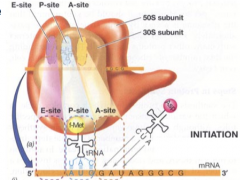
|
shinedalgarno mRNA sequence aligns with 165 rRNA of ribosome. shinodalgarno mRNA sequcne aligns with 16S rRNA of ribosome. mRNA with ribosome aligns with 16S, attaches at psite, and begins initiation |
|
|
translation: elongation hpic |
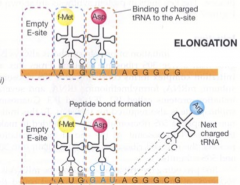
|
1) tRNA+AA binds to A-site (requires GTP) 2) peptide bond joins amino acids, (catalyzed by 23S rRNA) 3) ribosome moves 1 codon along mRNA 4) empty tRNA moves from P to E |
|
|
Translation: Termination |
-initiation: methionine coupled together with formyl group engages ribosomes at P-site -Elongation: bring another tRNA molecule with its corresponding Anti-codon, peptide bond formed tween two amino acids -Termination: takes place at any 3 stop codons, -UAA, -UAG, -UGA, (no tRNAs that correspond to these) -release factors, cleavage, release polypeptide |
|
|
|
Mutations and their chemical basis (hnotes) |
•Mutations –stable, heritable changes in nucleotide sequence relative to wild-type –may or may not effect phenotype •Wild-type strains possess the typical or representative characteristics of a species •Mutant strains have mutations - stable, heritable changes in nucleotide sequence relative to wild-type strain |
Although mutations may or may not effect phenotype, they are often classified in terms of their effect on phenotype ... •Morphological - change colonial or cellular morphology •Lethal - kill •Conditional - expressed only under certain conditions (e.g., high temperature) |
|
|
How Mutations Arise hnotes |
•Spontaneously –absence of added agents (errors in DNA replication) –Transitions, Transversions •However, DNA replication errors are rare – DNA pol works at rate of 1000 nucleotides per second •but incorporates incorrect base one in a billion •has proofreading activity |
•Induced - after mutagen exposure – chemical or physical agents - damage, alter DNA Examples: -UV light(generates thymine dimers) -Base analogs- resemble bases, cause mis-pairing to occur, ex) 5-bromouracil, T-analog that pairs with G -intercalating agents wedge themselves between certain segments
|
|
|
Mutations in protein-coding genes can affect protein structure in a variety of ways. The most common types of such mutations are: |
-missense: single base substitution, changes codon for one amino acid into codon for another amino acid -nonsense: converts a sense codon to stop (disables it) -frameshift: insertion or deletion of one or two base pairs in coding region of a gene |
|
|
|
More Types of Mutations (hnotes) |
-Auxotrophs: mutations in biosynthetic pathways. (auxo- refers to the mutant strain) cant make the product of the pathway. requires product in media. (ex: lysine auxotroph cannot make lysine) labeled lys- - |
replica plating is used to identify auxotrophs. |
|
|
Microbes are equipped with a variety of molecular tools that repair DNA damage, let’s consider two. |
-"light" repair: photoreactivation, light activated photolyase, binds and cuts thymine dimer, fixing it. -"dark" repair: nucleotide excision repair. UvrABC endonuclease removes section of damaged nucleotides. DNA polymerase I fills, and ligase joins. |
|
|
|
Creating Genetic Variability |
•Mutation (all domains) –new alleles, new phenotypes •Vertical gene transfer (Eukarya) –sexual reproduction –new combinations of genes when gametes from parents fuse •Horizontal gene transfer (Bacteria, Archaea)–transfer from one mature, independent organism to another |
|
|
|
Gene Transfer by Transformation |
-discovered by Fred Griffith -Uptake of free DNA from environment. -Competent cell: a cell that can take naturally take up DNA from the environment via transformation • Only a few bacteria are known to be naturally competent: •Gram +Streptococcus, Bacillus •Gram- Haemophilus, Neisseria |
|
|
|
Membrane-bound protein complexes bring DNA into the cell. How is DNA changed during this process? |
Fig16.26 gram positive vs gram negative gram positive: no outer membrane, needs additional proteins to allow DNA to enter cell gram negative: note* as DNA comes through barrier membrane, becomes unwounded by nuclease that is in membrane converst DS into SS |
|
|
|
Artificial Transformation |
•In lab with bacteria not naturally competent (e.g.,E. coli) •Critical step in cloning •Two techniques: -Calcium chloride: makes cells more permeable -electroporation: pulses high voltage, temporary holes- cell wall, plasma membrane |
|
|
|
DNA Transformation |
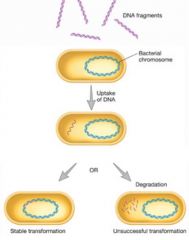
|
|
|
|
Gene transfer by conjugation hpic |
-DNA transfer by direct cell-to-cell contact -requires pill, and plasmids is commonly what is transmitted -major mode of spreading antibiotic resistance genes between bacteria |
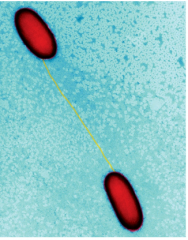
|
|
|
Plasmids |
•double-stranded, circular DNA •extrachromosomal •carry genes that confer advantage •can be transferred by conjugation • are replicons - have their own ori
-episomes- plasmids that exist with or without integrating into the chromosome. |
|
|
|
Example of conjugative plasmid (hpic) |
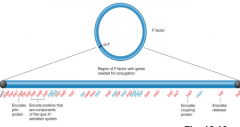
The F (Fertility) Factor of Escherichia coli is a well-studied example of a conjugative plasmid |
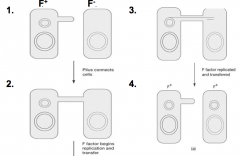
|
|
|
The F Factor is an Episome hpic |
-can integrate into chromosome -Cell now designated Hfr (high frequency of recombination) -Can transfer F factor and part of its own chromosome into F-cell |
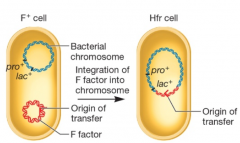
|
|
|
Transposition |
•Pieces of DNA move, insert different sites in genome •Transposable elements - called“jumping genes” –Transposons –Insertion sequences •Insert, turn genes on or off |
|

