![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
50 Cards in this Set
- Front
- Back
- 3rd side (hint)
|
What are telomeres? What is the sequence in humans?
|
-DNA sequence at the ends of linear chromosomes.
-They are composed of a simple DNA sequence (e.g., TTGGGG in the protozoan Tetrahymena .. TTAGGG in humans) repeated many times. How many times might the sequence be repeated in mammals? |
-In mammals the unit sequence may be repeated over 1500 times.
|
|
|
What cell lines have detectable levels of telomerase activity?
|
– germ lines and stem cells.
--Most somatic cells lack this activity, and the telomeres become shorter and shorter as the cell proliferates. |
|
|
|
Most somatic cells lack telomerase activity, and the telomeres become shorter and shorter as the cell proliferates. How many cell divisions can somatic cells undergo? Why is that an advantage to somatic cells?
|
-It limits somatic cells to 20-60 divisions.
-This is good because the more you replicate DNA the greater the chances are that you accumulate mutations or something may be altered in the DNA sequence. -If the sequence of the somatic cell becomes changed and it keeps on proliferating, it can evolve into something that doesn’t cooperate effectively with other cells (whomp whomp). -Because there was this problem in replicating chromosomes completely we have the telomere sequence. In somatic cells we don’t have telomerase activity so the lengths of the telomeres of somatic cells indicates the cells age. - A big example of this is??? |
TUMORIGENESIS!
-A cell can generate into a tumor cell, into a cancer cell, and then cause problems. -There is research that says, perhaps if introduce telomerase into cells like skin cells then you can reverse the aging process. -He is reserved about that type of research, doesn’t know if it is possible. |
|
|
What happens when telomeres shorten to a particular length?
|
-It’s like a clock – as it gets shorter, something kicks in when the length becomes a particular length and changes in gene expression, like entering into senescence (inability to replicate further, to license the origins of replication kick in).
-This helps to enforce the process by which we have these billions of somatic cells that are aiding the germ line cells to be immortal. -These somatic cells do not get out of line, they collaborate effectively, and when then become a particular age, if they accumulate particular mutations, for example, there can also be programmed cell death. Why might this be a good thing for the organism? |
-What we DON’T want is individual selection of a particular cell which, if it gains selective advantage over other cells, it’s good for that one cell in terms of individual selection but it is not good for the GROUP.
-This type of system probably evolved because it’s a type of GROUP selection – what is good for the GROUP of cells as a whole organism. |
|
|
Why are telomeres necessary?
|
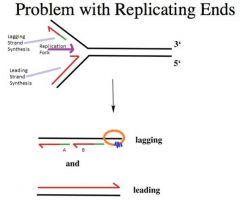
-They are required because DNA replication to the very end of linear chromosomes poses a problem.
-Arises from the property of DNA replication -DNA replication uses DNA polymerases -->All the known DNA polymerases require a preexisting primer for it to catalyze DNA synthesis -What happens is that it becomes impossible to completely replicate a linear chromosome Eg. -Looking at a replication fork going from left to right, note leading and lagging strand synthesis as indicated -In green is the RNA primer and Red is the newly synthesized DNA -Completely replicating the leading strand template is no problem because the leading strand polymerase can go all the way to the end of the molecule -The problem is with the LAGGING strand because lagging strand synthesis occurs in the direction away from the fork -You can do lagging strand synthesis almost all the end to the template, but there is always a little portion of the lagging strand template that remains unreplicated (orange circle) -When we remove the RNA primers (A), we degrade the RNA and fill in the gap created by removing the RNA by DNA polymerase activity using the adjacent Okazaki fragment (B) as the primer for gap DNA synthesis -Even if you were able to catalyze an RNA synthesis right at the very end of the lagging strand (blue squiggle) template, the question becomes how do you remove that RNA primer? -->There is no adjacent Okazaki fragment to fill in that gap in genetic information. When that RNA gets removed, we’d still be left with the same problem. There is no way to completely replicate the lagging strand -Even though the leading strand template is completely replicated, the same problem with the lagging strand exists at the other end of the chromosome as well. |
|
|
|
What happens when we replicate with a DNA polymerase that requires an RNA primer?
|
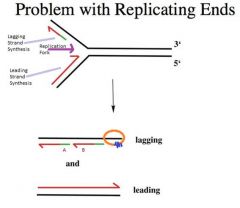
-If we were to replicate with a DNA polymerase that requires an RNA primer, what happens is that from generation to generation with every replication cycle the chromosome gets shorter and shorter.
-This is what happens in the vast majority of our somatic cells. Our chromosomes are becoming shorter and shorter |
|
|
|
Germ line cells and stem cells must propagate its DNA and give rise to many other cells. How are they able to do this if chromosomes get shorter every time the cell divides?
|
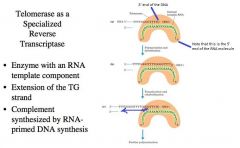
-Germ line cells and stem cells must propagate its DNA and give rise to many other cells.
-We have an enzyme called telomerase. -Telomerase is a specialized type of reverse transcriptase where there is an RNA that acts as a cofactor for this enzyme. -The RNA (depicted in green) is in the protein (orange). -Telomerase adds on the TTGGGG sequence to the ends of the telomere sequence, thus expanding the telomere sequence -In germ line cells and stem cells, the RNA cofactor in telomerase hybridizes to the 3’ end of the DNA (noted w/ blue arrow .. Also note the 5’ end of the RNA molecule) -We use the RNA as a template and we extend and copy the template as shown in B -The telomerase sequence translocates and another cycle of DNA synthesis using the RNA template can be catalyzed -We extend upon the 3’ end of the telomere sequences, expand using the RNA as a template. Once we get rid of the telomere sequence we can do an RNA primer (blue squiggle) synthesis and extend (line + arrow) and maintain the lengths of the telomere |
|
|
|
What type of enzyme is telomerase?
|
-Reverse transcriptase: DNA polymerase that can use RNA as a template
|
|
|
|
What can reverse transcriptases use as a template? What can DNA polymerases use as a template for DNA synthesis?
|
-Most RT we know of can use RNA OR DNA as a template
-Most of our known DNA polymerases cannot use an RNA at all as a template for DNA synthesis |
|
|
|
What can viral genomes be composed of?
|
-There are DNA and RNA genomes
-The duplex DNA genome is ideal for easy replication and maintenance. -+ and – RNA genomes -single and double stranded -segmented genomes Viral genomes are: -Fairly flexible – not just double stranded DNA! -It can also be single stranded DNA, double stranded DNA, as well as RNA genomes. -RNA genomes have an advantage - Negative RNA has an intrinsic plasticity about it that DNA has less of. -When we have single stranded RNA it can be + or – polarity. |
|
|
|
What is the difference between + and – RNA?
|
+ polarity means that the polarity of the genome has the same polarity as the principle messenger RNA.
An RNA virus that has a + polarity, that RNA can enter into the cell and be used immediately as mRNA. Something with – polarity, like influenza virus which has an RNA genome of negative polarity, that RNA has to be copied by an RNA dependent RNA polymerase to + RNA before it can be used as mRNA. |
|
|
|
Name some characteristics of larger viral genomes.
|
The larger genomes tend to be DNA, duplex, and very large.
They may encode their own replication apparatus. |
|
|
|
Name some characteristics of smaller viral genomes.
|
The smaller genomes have the option of being single stranded, probably because of the fact that the smaller the genome the more plasticity and errors and damage that can be tolerated.
Eg. influenza virus. Tend to be RNA genomes. |
|
|
|
The most rapidly evolving genomes tend to be composed of?
|
The most rapidly evolving genomes tend to be RNA.
Eg. AIDS |
|
|
|
In what ways can viruses be dependent on host cells? What can influence this dependency?
|
-The genomes of viruses contain sufficient information to exploit the host cell apparatus for its own replication and packaging into viral particles.
-The size of a viral genome can vary greatly, from 3 to 300 kb. -Thus, The virus is dependent on the host to different degrees -Because these are viruses, they are all dependent on the host machinery to translate mRNA. -They need the cellular compartment provided by the host. -Viral replication systems must therefore be economically designed, making the most of limited size of the genome. -Some viruses encode many of their own replication enzymes while others rely heavily on the host replication apparatus, sometimes even dependent on other helper viruses for productive infection. -They are dependent to different degrees in that some viruses, like the larger herpes simplex viruses, encode their own initiator protein for initiating DNA replication, their own DNA polymerases, even their own enzymes for making their own nucleotides through salvaging mechanisms -Smaller genomes are heavily dependent on cellular replication apparatus because its genome is too small to encode its own DNA polymerase and its own nucleotide synthesizing enzymes. Eg. papillomavirus – E6 and E7 bind to p53 and Rb |
|
|
|
Describe the different strategies of the herpes simplex virus and the papillomavirus. Why might these strategies differ?
|
-They are dependent on the host genome to difference degrees in that some viruses, like the herpes simplex viruses, encode their own initiator protein for initiating DNA replication, their own DNA polymerases, even their own enzymes for making their own nucleotides through salvaging mechanisms whereas a very small virus, like the papillomavirus, which is largely dependent on the cellular replication apparatus because its genome is too small to encode its own DNA polymerase and its own nucleotide synthesizing enzymes.
-What that means is that when the herpes simplex virus takes over the cells to replicate its own DNA, it starts expressing its own DNA polymerases and comes up with a replication system that is independent of the host to promote high amounts of its own replication whereas something smaller like the papillomavirus will do things to encourage the cell to enter the S phase of the cell cycle. Eg. Papillomavirus produces proteins E6 and E7 that bind to gatekeeper tumor suppressors called p53 and Rb. P53 and Rb are tumor suppressors that put a break on the cell cycle. They regulate entry into the cell cycle. E6 and E7 bind to p53 and Rb and makes the cell enter into the cell cycle and makes the host proliferate incessantly -->What this does is to promote the cell to be in S phase producing lots of nucleotides and producing a lot of replication enzymes for the cell to promote its own replication. -->In that process, the papillomavirus has lots of materials it can commandeer to propagate its own genome for its own replication. -->It serves the papillomavirus to have the cell persistently dividing, and that’s how papillomavirus can cause such things as warts or cancer. *Papillomavirus is the causative agent of cervical cancer. |
|
|
|
How do the sizes of viral genomes compare to those of cellular genomes? What implications does this have?
|
-In all cases, whether it is a large or small viral genome, the genome is small in comparison to the cellular genome and thus a higher mutation rate can be tolerated – a little bit less with the larger viral genomes like herpes simplex virus, but a lot more mutations can be tolerated by viruses like the influenza virus with a smaller genome.
-The duplex DNA genome is ideal for easy replication and maintenance. -However, viral genomes are relatively small and have a significantly less chance of being damaged. -Also, in a productive viral infection, many viral particles are released. |
|
|
|
Does the system that replicates the viral genome need to be as accurate as the cellular replication system? Why or why not? Is an accurate system an advantage to a virus?
|
The system that replicates the viral genome does not necessarily need to be as accurate as the cellular replication system.
In fact, viral genomes may be more likely to prosper with an error-prone system that would increase its genetic diversity. |
|
|
|
Describe the influenza virus genome.
|
-Influenza virus has its genome in multiple, single-stranded RNA copies.
-The RNA is said to be a negative strand--opposite to the polarity of mRNA necessary to translate the viral gene products What is the advantage of having an RNA genome??? |
-RNA has the advantage of having the plasticity that allows the RNA genome to change very rapidly.
- It is important to note that you can have a very rare even that somehow gives the RNA genome a selective advantage in a particular environment. -This can allow the virus to propagate and become the dominant virus that is present in a particular population, perhaps even causing an epidemic. |
|
|
What is a replicase? Where does it act?
|
RNA-dependent RNA polymerase (replicase)
-On a linear RNA genome, the replicase can initiate RNA synthesis starting at the 3' end. -There is no mechanism for repairing errors, which take place once in 104 nucleotides incorporated. -The genome size of RNA viruses are necessarily small. |
|
|
|
What do RNA viruses require for their replication? Does this differ between + and – strand RNA viruses?
|
***RNA-dependent RNA polymerase***
-With the exception of the retroviruses, RNA viruses require an RNA-dependent RNA polymerase (replicase) for their replication. -Most of the RNA viruses have to encode its own RNA dependent RNA polymerase because a cell doesn’t have an effective system for replicating the RNA genome from RNA viruses -The negative-strand RNA viruses must have the replicase packaged in the virion with its genome whereas the positive-strand RNA genome may be translated to synthesize the replicase Eg. influenza virus needs an RNA-dependent RNA polymerase that converts the RNA genome of negative polarity to positive RNA that can be translated into viral proteins -There is no mechanism for repairing errors, which take place once in 104 nucleotides incorporated. -The genome size of RNA viruses are necessarily small. -The high rate of error as well as the segmented arrangement of the influenza genome helps it to evolve into new strains that can evade host immune system. |
|
|
|
What do herpes simplex virus and papolloma virus need to replicate?
|
DNA polymerase
Does herpes make its own? Papolloma virus? |
In the case of herpes simplex virus it encodes its own DNA polymerase
Papolloma virus is more dependent on the cellular replication machinery so it does not encode its own machinery |
|
|
What is reverse transcriptase? How was it discovered?
|
-Reverse Transcriptase has all the characteristics of a DNA polymerase except for the fact that it can use not only DNA but also RNA as a template
-This was discovered first in association with retroviruses Retroviruses -->RNA viruses that replicated via an DNA intermediate. -That DNA intermediate can also be considered as a transposable element that can transpose via an RNA intermediate and that intermediate can be a viral genome. -We have many different kinds of retro-elements, transposable elements that transpose via an RNA intermediate. -Some are like the retroviruses and package their RNA intermediate into viruses or virus like particles. -There are others, like L1, the dominant transposable element in humans, which is not an LTR type retrovirus. (We are not looking at L1 elements, rather we’re looking at LTR type elements as represented by retroviruses. ) |
|
|
|
What are retroelements?
|
Retroelements are endogenous components of eukaryotic genomes that are able to amplify to new locations in the genome through an RNA intermediate.
|
|
|
|
What are LTR’s?
|
LTRs: They represent a class of transposable elements which include non-viral elements such as the yeast Ty elements.
-When the Ty element transposition mechanism was discovered, even those these Ty elements are not virus it was discovered they had a gene structure analgous to retroviruses, and even the RNA intermediate was packaged into virus like particles even though they weren’t actually viruses. This was when the retrovirus was identified as a transposable element that moved via an RNA intermediate. Wiki says: The LTRs are partially transcribed into an RNA intermediate, followed by reverse transcription into complementary DNA (cDNA) and ultimately dsDNA (double-stranded DNA) with full LTRs. The LTRs then mediate integration of the retroviral DNA via an LTR specific integrase into another region of the host chromosome. |
|
|
|
What are retroviruses?
|
Retroviruses
-->RNA viruses that replicated via an DNA intermediate. What can that DNA intermediate also be considered as? |
-That DNA intermediate can also be considered as a transposable element that can transpose via an RNA intermediate and that intermediate can be a viral genome.
-We have many different kinds of retro-elements, transposable elements that transpose via an RNA intermediate. -Some are like the retroviruses and package their RNA intermediate into viruses or virus like particles. -There are others, like L1, the dominant transposable element in humans, which is not an LTR type retrovirus. -We are not looking at L1 elements, rather we’re looking at LTR type elements as represented by retroviruses. |
|
|
What happens when retroviruses enter a cell?
|
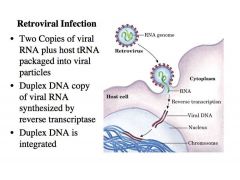
-Retrovirus is a membrane bound nucleocapsid that has ****Two copies of viral RNA genome**** plus host tRNA packaged into viral particles
-When the RNA is injected into the cell, the reverse transcriptase that is packaged with the RNA will eventually convert that RNA into a Duplex DNA copy of viral RNA -This Duplex DNA is integrated into a random site of the host chromosome by an enzyme called INTEGRASE -Integrase has an activity that is virtually identical to the transposase from lecture 12 – it binds to the ends of the viral provirus DNA and it integrates into a random site of the host chromosome -Once the virus is integrated into the host chromosomes this piece of DNA gets transcribed, and we make viral RNA. The viral RNA is packaged into these virus like particles and released as virus into the environment to infect another cell. |
|
|
|
Define: provirus:
|
-The enzyme reverse transcriptase is an enzyme that can synthesize DNA using an RNA or DNA molecule as template.
-This enzyme initiates DNA synthesis on the retroviral RNA using the tRNA primer, which binds to a specific site on the template. -Once the negative strand of DNA is made, reverse transcriptase synthesizes the positive strand. -The resulting duplex DNA has a terminal redundancy consisting of U3-R-U5, which is called the long terminal repeat (LTR). -Integrase encoded by the retrovirus catalyzes the integration of the duplex DNA molecule into the host chromosome. **The integrated retroviral DNA is called the provirus.** |
|
|
|
What integrates the retroviral DNA into the host genome? Is this integration targeted or random? How often is it successful?
|
-Integrase, which is encoded by the retrovirus, catalyzes the integration of the duplex DNA molecule into the host chromosome.
-Integration is at random sites on the chromosome, a successful infection producing 1 to 10 proviruses per cell. |
|
|
|
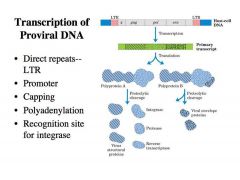
Describe the structure of the PROVIRUS !
|
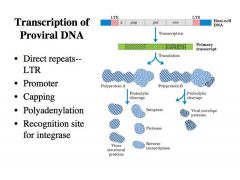
-This viral DNA that is integrated into the chromosomes is called the PROVIRUS.
-It has the following structure: a. At the left and right ends are DIRECT repeats called LTRs or LONG TERMINAL REPEATS What is the function of the LTRs? What are gag pol and env? |
It has several different functions:
1. It provides the promoter for transcription to make the primary transcript from the provirus DNA -The U3 region of the LTR carries a promoter, allowing the synthesis of retroviral RNA. -This RNA is translated for viral gene expression and virion production, and it is also the genome packaged into virions. -The details of translation are not so important, just note that all these viral gene products are translated as one giant polyprotein and these polyproteins are cleaved by proteolytic cleavage to give rise to the individual products. 2. Has sequences that direct the capping and the polyadenylation as transcription reaches the end of the proviorus DNA -This RNA looks in every way like the regular mRNA --> Cap +polyA tail 3. Provides the ends to which the integrase binds. LTR provides the binding sites for integrase to bind and promote integration Gag pol and env are the specific proteins necessary to assemble the virus. Pol is especially important, because this is the region that encodes the integrase and the reverse transcriptase. |
|
|
Describe the structure of the duplex DNA made from the viral RNA.
|
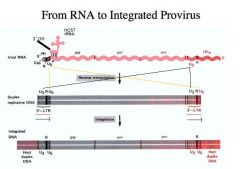
1. Look at the structure of the mRNA --> Here is the mRNA of the retrovirus
-Note that it has the cap and poly A tail -The sequence adjacent to the cap is called the R sequence -It’s called R because it’s a REPEAT that’s found right by the cap and right adjacent to the poly A tail -RNA has a terminal redundancy of 13 to 250 bases (R). -There is a U5 sequence -This means there is a “unique” sequence at the 5’ end -At the 5' end, a unique sequence of 80 to 200 bases (U5) flanks the R sequence. -Right next to U5 is where the tRNA is bound. This is the HOST tRNA. Note the orientation of the 3’ OH end of the tRNA -There is also a U3 sequence -This means there is a “unique” sequence at the 3’ end -At the 3' end, a unique sequence of 170 to 1200 bases (U3) flanks the R sequence. THEN reverse transcription does is to make a COPY of the RNA. -Notice, however, that it is not the R sequence that is repeated here, but the sequence with the long terminal repeat. -The long terminal repeat consists of: 1. U3 sequence 2. R sequence 3. U5 sequence -That is, we combine sequences from both ends of the viral RNA together with the common R sequence at the left and right ends to make the LTRs. The same orientation is present at the left and right ends. It is this duplex DNA which is mostly identical to the viral RNA but not quite because the LTR is different that gets integrated into the host genome. |
|
|
|
What does the long terminal repeat consist of?
|
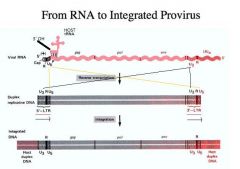
The long terminal repeat consists of:
1. U3 sequence 2. R sequence 3. U5 sequence -That is, we combine sequences from both ends of the viral RNA together with the common R sequence at the left and right ends to make the LTRs. -The same orientation is present at the left and right ends. - It is this duplex DNA which is mostly identical to the viral RNA but not quite because the LTR is different that gets integrated into the host genome. |
|
|
|
What is Rnase H? What does it do?
|
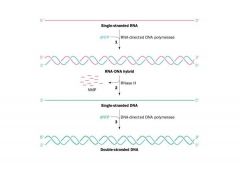
DISCLAIMER:
*This slide simplifies the process by which we get transcription of the single stranded RNA to duplex DNA *CAUTION : something is wrong with this slide!!! It has a BAD ERROR THAT CONFUSES THE ISSUE! *He’s going to go through it anyway!!!!! HIROSHI DANGERRRR NAKAI!! -Here is the RNA genome and here we see the reverse transcriptase -The red is the RNA and the blue is the DNA and reverse transcriptase has made a complementary strand of the RNA -Reverse transcriptase has a very potent activity called Rnase H. Rnase H is an RNAse, meaning it degrades RNA that is specifically in an RNA-DNA duplex. -As soon as reverse transcriptase copies the RNA and RNA becomes part of this RNA-DNA duplex, it becomes susceptible to degradation and its degraded away. -Since reverse transcriptase can replicate both a template that is RNA as well as one that is DNA, it synthesizes a complementary strand of the DNA and then you get double stranded DNA that becomes integrated into the host chromosome. Psh, that didn’t seem so bad NAKAI. What is the (your?!) problem with this model? |
-Reverse transcriptase is a DNA polymerase and you need a primer to replicate a linear chromosome
-How do you completely replicate a linear template using a DNA polymerase??? It’s impossible by this mechanism! If you were to make an RNA primer that is depicted here in blue, if part of that were an RNA primer it would be degraded away and you wouldn’t be able to completely replicated the RNA genome. -The big clue into this comes from the fact that if that model from the previous slide was right, it should be the R sequence that is repeated at the left and right ends of the provirus DNA, not LTR. |
|
|
What does the actual mechanism of retroviral DNA synthesis look like?
|
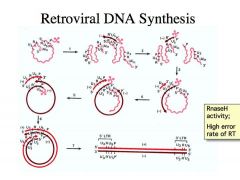
The reason why they simplify it like this is because the actual mechanism looks like THIS.
-It’s much more complicated and it’s a headache to learn, so we don’t have to memorize this mechanism. -If you get to the bottom of it, it has a simple lesson associated with it. -That is, if you want to replicate a linear molecule completely, you need a primer and you need to be able to do synthesis all the way to the end of the linear molecule and lose the priming site and not have any consequences. -We don’t need to know the whole thing, but we need to learn the lessons. -It is the tRNA that is the primer for DNA synthesis. The tRNA is bound very close to the 5’ end, so there is only a little bit of distance that RT can go if it uses the 3’ end of the tRNA as a primer for DNA synthesis -As soon as you do DNA synthesis, the RNA in the RNA-DNA hybrid becomes susceptible to degradation, and it is degraded almost immediately. -This creates an R sequence that’s complementary to the R’. R’ will now hybridize to the 2nd retroviral RNA. Now you get priming and DNA replication. Then you form the LTR sequence. -This mechanism is a way of completely replicating that end of the molecule. You have completely replicated the R and you have a redundant sequence because that redundancy helps the full RNA to be copied. ___ Soooo this is all we really need to remember, but for the sake of COMPLETENESS, herewego at the other end of the molecule(cue N’sync): -When we go to the other end of the molecule again now have another problem because when we get to the 5’ end we only have an R sequence and we need to have another LTR at that end! -When we degrade the RNA template on this newly synthesized single stranded DNA (orange arrow), one of the fragments of RNA from degrading the RNA template becomes a primer for DNA synthesis in the direction of the blue squiggle arrow -Again you make U3, R sequence and U5 from copying this region of the DNA template, the first complementary DNA strand that was synthesized. It collides with this tRNA. As soon as we get DNA synthesis into the tRNA region, the tRNA gets degraded away (red squiggle) by the Rnase H activity. This allows the P site to hybridize to the complementary P’ site on the DNA. -We THEN get DNA synthesis here (green arrow) and here (purple arrow) and now we lead to the replication of PROVIRUS DNA that has an LTR at the left and right ends This is why the retroviral genome is so dynamic ! "The key to this is not memorization." Instead, what are two lessons we can take away from this arduous example? What is really most important to take away from this complex mechanism?: |
1. Because of the fact that reverse transcriptase tends to be error prone and it doesn’t have a proofreading function associated with it and because the RNA tends to have a plastic quality to it such that if the cytosine residues become deaminated to uracil residues that situation is going to hold, and HIV reverse transcriptase is one of the most error prone DNA polymerases known. It makes a mistake about 1 in every 10,000 times. That is about how many errors you would expect to get if all the tautomeric transitions that the nucleotide template can undergo had an influence on nucleotide selection. There is minimal effort made by reverse transcriptase to discriminate the nucleotides. It just incorporates whatever fits into the active site. For that reason, there are always lots of mutations being introduced into the retrovirus genome
What’s most important is this complex mechanisms that requires two RNA viruses for the making of a complete provirus DNA. This becomes a mechanism for recombination between two retroviral strains. |
|
|
Say that you have two proviruses in a particular cell – one that confers resistance against chemotehrapeutic agent A and a second retrovirus that confers resistance against chemotherapeutic agent B.
Could both of these somehow end up in the same virus? How might this happen? Hint: the answer is not magic. |
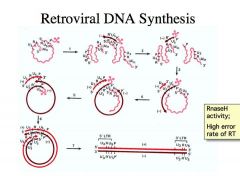
-You can make this happen because of the way retroviral DNA synthesis works – it requires two RNA viruses for the making of a complete provirus DNA. This becomes a mechanism for recombination between two retroviral strains.
-Imagine you have two proviruses in a particular cell – one that confers resistance against chemotherapeutic agent A, and a second retrovirus that confers resistance against chemotherapeutic agent B. Those mutations that lead to resistance to chemotherapeutic agents can arise fairly readily because of the high mutation rate afforded by the reverse transcriptase, but if you have to have two different RNA molecules for making a proviral DNA, that means two different RNA from two different retroviral strains can be packaged into the SAME virus particle!!! When it makes this provirus DNA, a rare event could potentially incorporate it into the same provirus, the allele in one position and the allele in another position, and thus you can get a retroviral strain that confers resistance to both chemotheraputic agent A and B. Because it has the selective advantage, even if it’s a rare event that virus will propagate and become a prominent virus in a particular population. |
|
|
|
How error prone is the HIV reverse transcriptase?
|
-Reverse transcriptase, like the replicase of RNA viruses, does not have a system for error correction.
-Thus, there is not only the relatively high error rate by the reverse transcriptase that synthesizes the proviral DNA, there is also the high error rate by the RNA polymerase that transcribes that DNA. -The HIV reverse transcriptase is especially error-prone. It has a misincorporation rate of 1 per 2000 to 4000 nucleotides polymerized. |
|
|
|
Does reverse transcriptase have an Rnase H activity?
|
-Reverse transcriptase also has an RNase H activity, a nuclease that cleaves RNA in RNA-DNA hybrids.
|
|
|
|
What is the role of retroviruses in converting proto-oncogenes to oncogenes?
|
-The retroviral genome sometimes will acquire host DNA, commonly a gene involved in cell growth.
-This protocogene if mutated can be converted to an oncogene. -The error-prone replication system of the retrovirus is a good way of producing oncogenes. -A cell infected with a retrovirus carrying an oncogene will become tumorigenic. What is a classic example? |
-A classic example is the avian sarcoma virus carrying a 60-kDa phosphoprotein that is a protein kinase.
-This viral oncogene (v-src) has a host counterpart (c-src). |
|
|
What is the strategy behind the use of anti-viral agents?
|
-Drugs which inhibit viral replication are commonly used to control viral infections (e.g., infections with HSV or HIV).
-The strategy is to find drugs that will effectively inhibit the viral replication apparatus with a minimal effect on the host replication system. Ie. Basic strategy: inhibit viral DNA synthesis but not that of the patient -For example, one strategy may take advantage of the simpler design of the viral replication apparatus, which may not discriminate nucleotide analogs as effectively as the more complex host replication apparatus. |
|
|
|
How can you treat HSV infections?
How does treatment work? |
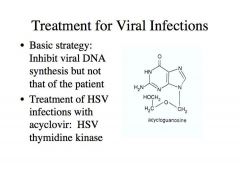
-One successful and effective treatment that is known is the treatment of HSV infections with acyclovir: HSV thymidine kinase
Acyclovir is a nucleoside analogue -The image shows acycloguanosine and what you can see is that it is the guanine base attached to what looks like the ribose moiety of a nucloeside but it’s lacking the 2’ and 3’ carbons -It has what would be considered the 5’ carbon shown here -It is a nucleoSide. If it’s not phosphorylated, it cannot be used as the precursor for DNA synthesis -In fact, our cells lack any enzymes to use this molecule and phosphorylate it into a form that will activate this into a nucleotide. -A herpes simplex viral genome has nucleotide salvaging enzymes! -HSV encodes something called THYMIDINE KINASE. It can take nucleosides like this and phosphorylate at this position, converting acycloguanosine into a monophosphate. -Once this is a monophosphate, cellular enzymes can convert this into a diphosphate and a triphosphate, and then we have an acyclovir 5’ triphosphate that can be used for DNA synthesis by HSV DNA polymerase. What happens to this molecule then in a HSV infected cell? What effect does it have on a cell that is not infected with HSV? |
-If you have a HSV infected cell, it has the THYMIDINE KINASE to convert this into a substrate for DNA synthesis, it gets incorporated by the HSV DNA polymerase. When it gets incorporated, it is a CHAIN TERMINATOR because it lacks a 3’ OH to continue on with the polymerization process
-As soon as this gets incorporated, DNA synthesis stops. -On the other hand, the host thymidine kinase cannot use acyclovir as substrate, giving the drug its specificity for HSV-infected cells. -This is a way to stunt the replication of HSV while not affecting replication in normal replicating human cells because it cannot activate this acycloguanosine. |
|
|
How do we treat AIDS?
|
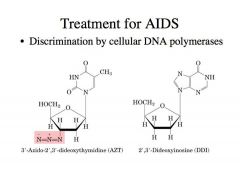
-Treatment of HIV infections with 3'-azido-2',3'-dideoxythymidine (AZT) and 2',3'-dideoxyinosine (DDI).
-There are nucleoside analogues -AZT has an azido group, so it becomes a chain terminator -->If this is activated as a nucleotide and incorporated, this azido group prevents further polymerization events -The same is true for dideoxyinosine. DDI is an example of a newer drug with a mechanism of action similar to AZT. -->The 3’ deoxygroup prevents further polymerization once this is incorporated -Cellular cells have kinases that can phosphorylate AZT to AZT triphosphate. -Those triphosphates are used by the AIDS virus reverse transcriptase to replicate and make the provirus DNA -AZT effectively inhibits the reverse transcriptase because this enzyme has a higher affinity for AZT triphosphate than for dTTP (i.e., the Km for AZT triphosphate is lower). -Incorporation of AZT by reverse transcriptase results in chain termination. -Thus, we prevent the AIDS virus from creating the provirus that will integrate into the host chromosome. Why doesn’t this affect the host polymerase as much? |
-Host polymerases are more discriminatory. IT will take these triphosphate versions of these but it wont recognize them as nucleotides very efficiently because of the careful nucleotide selection.
-AZT does not affect the cellular DNA polymerase, which is 100-fold less sensitive to AZT and has a much higher affinity for dTTP. -Even if they are incorporated, cellular DNA polymerase has the proofreading function the 3’5’ exo activity that can back up, remove incorporated nucleotide analogues like this such that DNA synthesis can be resumed. -Reverse transcriptase lacks such a proofreading function so once this is incorporated into its DNA, that’s the end of that particular DNA template |
|
|
At what concentration is AZT effective in inhibiting HIV reverse transcriptase?
Is AZT toxic to T cells? Bone marrow cells? |
AZT present at 1 to 5 µM is effective in inhibiting HIV reverse transcriptase but is not toxic to T cells.
Unfortunately, it appears to affect bone marrow cells, causing anemia. |
|
|
|
Name two uses of Viral Vectors in Gene Therapy. Give some examples too!
|
A. Treatment of human disease by gene transfer.
1. Correction of single gene defects by introducing the functional gene. a) The first human gene therapy trial involved the transfer of the ADA (adenosine deaminase) gene into lymphocytes (begun September 1990). -->The principle is as follows: If you have an inherited disease such as SCID that results from ADA deficiency, you use viruses as vectors to introduce the ADA gene into the lymphocyte b) Treatment of the Lesch-Nyhan syndrome by introduction of the gene encoding hypoxanthine-guanine phosphoribosyltransferase. -In more recent years, viruses as vectors have been frowned upon because they have caused a considerable amount of problems in patients. 2. Gene therapy in the treatment of acquired diseases. -If you have an acquired disease such as cancer, you make the lymphocytes of the patients more conducive to killing their own cancer cells by introducing genes such as the gene for TNF into the lymphocytes so that these lymphocytes become super cancer killing cells. -For example, transfer of genes to immune cells of the patient to stimulate destruction of tumor cells by the host immune system. -The tumor necrosis factor (TNF) gene has been transferred into tumor-infiltrating lymphocytes, which are reintroduced into mice with tumors. These modified lymphocytes migrate to the tumor and produce a high local concentration of TNF, which stimulate host immune response against tumor cells. |
|
|
|
Name the methods of introducing DNA into cells.
|
1. Injection.
-->Microinjection DNA into individual cells may cause mechanical damage to cells. This also a time-consuming process requiring specialized skills and equipment. 2. Transfection. -->For example, use of calcium phosphate or entrapment of DNA within liposomes to pass the DNA through the cellular membrane. 3. Infection. -->Viruses have evolved strategies for efficient introduction of genetic information into cells. Viruses are therefore effective vehicles of engineered DNA for gene transfer. Moreover, the specificity of the viruses for cell types may be exploited. A major problem is to avoid any cytotoxicity or diseases caused by these viruses. *nakai rushed through the end of this lecture and didn't mention this in class, but it's in the syllabus. I guess it comes down to how much you thirst for knowledge... |
|
|
|
What are the advantages of Viral Vectors?
|
1. High efficiency of gene delivery
2. Retroviral vectors allow the introduced gene to be stably integrated into the cellular chromosome. Full length copies of the gene are integrated. 3.Viral vectors are defective for replication so they do not disseminate |
|
|
|
Principal viral vectors with potential applications for human gene therapy.
|
1.Retrovirus.
-->The gene carried on the viral vector can be integrated into the cell genome. 2. Adenovirus. -->This virus has a number of useful attributes such as the ability to infect cells that are refractory to retrovirus infection; however, viral genome does not normally integrate into the host cell genome. 3. Adeno-associated virus. -->This virus has the most limited capacity for foreign DNA (4 kb), and its biology is not well understood. -->Its genome does integrate at a specific site on human chromosome 19. 4. Herpes Simplex Virus. -->This vector has a very large capacity for foreign DNA (30 kb). -->The virus can infect nondividing target cells such as neurons. *nakai rushed through the end of this lecture and didn't mention this in class, but it's in the syllabus. I guess it comes down to how much you thirst for knowledge |
|
|
|
What are the targets for Gene Therapy ?
|
-he said he wasn’t going through these, that this was not important for our purposes, but one thing he did say was that genes cannot be introduced or manipulated in FERTILIZED cells for ethical reasons because what we get is a transgenic animal when we do that – if we introduce a new gene into a fertilized cell that enters into the germ line and the engineered gene can be propogated into offspring.
-Transgenic animals have been of utmost importance in biomedical research as a mechanism of discovering what happens when we knock out a particular gene, for example, or what effect a mutation in certain genes might have. ____ The slide said: -Undifferentiated cells -->In practice, not ideal targets for gene therapy -->Investigators initially assumed that hematopoetic stem cells would be ideal targets for initial trials for gene therapy. Many diseases are due to single-gene defects in cell types derived from stem cells. However, there have been problems in efficiently transferring DNA to these cells. In addition, there have been problems getting expression of the transferred gene, a common problem in gene transfer methodology. -Differentiated somatic cells -->Eg. low density lipoprotein receptor gene introduced to hepatocytes -Genes cannot be introduced or manipulated in fertilized cells for ethical reasons -->Transgenic animals in biomedical research |
|
|
|
How does a retroviral vector work?
|
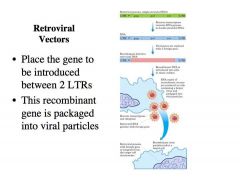
-WE don’t need to learn this, but he wanted to give us a brief idea as to how a retroviral vector works.
-Here is the retroviral vector at the top in green, and the viral proteins (gag, pol, and env) -This is also present in the DNA (blue) The idea is to replace all the viral genes with a gene of interest that you want to introduce into a particular cell type, lets say adenosine deaminase gene in pink. WE can replace that into the retroviral vector -Note the psi sequence at the 5’ end of the RNA that allows the RNA to be recognized as viral RNA -It is then packaged into viral particles. -What we do is to take this retroviral vector with a particular gene of interest introduced inside in place of the viral genes and we place it in a packaging cell line. -That packaging cell line has a retroviral genome that does express the viral proteins, but the retroviral genome here has an inactivated psi so its RNA is not actually packaged into virus particles, but the translated proteins are used to make virions and in these virions we package specifically the RNA that comes from the recombinant retroviral DNA. -Those viruses are then used to introduce a particular gene into a particular cell type. |
|
|
|
What are the disadvantages of Retroviral Vectors?
|
1. Cell types that can be infected are limited
--> This is not as important as fact 2 2. Outbreak of replication-competent retrovirus is possible -We saw how retroviruses can readily recombine by the mechanism that he spent so much time on .. -A rare event is all that is necessary to make a replication-competent retrovirus, which can then cause disease 3. Retrovirus integration is random and can be mutagenic |
|
|
|
Briefly describe gene therapy for SCID.
|
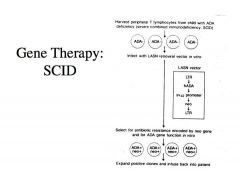
-ADA deficiency = SCID
-Gene therapy was used with limited success in 1990 -They took T lymphocytes from a child with ADA deficiency -Harvested ADA deficient cells and infected them with virus that has a functional human ADA gene that replaced the viral gene as well a neomycin resistance gene. -The neomycin resistance gene is there so that once the cells become infected with virus you can select for them by growing them in the presence of an antibiotic called gentamycin -We can now expand the lymphocytes from the child and the ADA gene allows the lymphocytes to expand since restoring ADA allows DNA replication to take place more efficiently -Then, this is infused back into the patient -This restored a considerable amount of immune function into the patient -At the time, it was a very encouraging result The only problem? It is very difficult to get the full repertoire of lymphocytes by just harvesting and trying to infect --?That’s why it had limited success BUT when you have a particular disease and you have a deficiency in one particular gene product, these diseases have the potential for some type of gene therapy in which we introduce a functional gene into these cell types |
|

