![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
61 Cards in this Set
- Front
- Back
|
What is the Michaelis Menten Equation? |
v = (Vmax * [S]) / (Km + [S]) |
|
|
What is the equation for Km? |
Km = (K-1 + Kcat)/K1 |
|
|
What is the definition of Kcat? |
Kcat =(K2 * K3)/ (K2 + K3) |
|
|
What does Km stand for phisiologically? |
Km is the substrate concentration where v is 1/2 of Vmax |
|
|
When does Vmax occur? |
When all of the enzyme is in use |
|
|
What is the rule about the lowest concentration of substrate in a rate experiment |
At the lowest levels, substrate concentration shouldn't be depleted by more than 10% during the course of the experiment |
|
|
how are enzyme catalyzed reaction kinetics often studied? |
by varying the amount of substrate and measuring product formation |
|
|
what is the rate at low substrate concentrations? [S] << Km |
it's first order (that is to say that doubling the substrate doubles the rate) |
|
|
what is the rate order at high substrate concentrations |
Zero order, doubling the substrate won't change the rate |
|
|
what is the rate order of an enzymatic reaction is near Km? |
1/2 order, doubling substrate increases the rate by 50% |
|
|
What are the axes of a lineweaver burke plot? |
1/v and 1/[S] |
|
|
What is the y-intercept of a lineweaver burke plot? |
1/Vmax |
|
|
For a lineweaver burke plot, what is the x-intercept? |
i/Km |
|
|
what are the axes for eadie hofstee plots? |
V & v/[S] |
|
|
How can you determine Km and Vmax from an Easie-Hofstee plto? |
Slope = -Km y-intercept = Vmax |
|
|
What does positive cooperativity look like on an Eadie Hofstead Plot? |
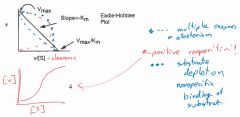
|
|
|
What are the three major types of non-Michaelis Menten kinetics that reciprocal plots help detect? |
multiple enzyme activity non specific binding allosteric behavior |
|
|
Who (in what units) Is Km reported? |
In terms of concentration (Molar, nM, micromolar, etc.) |
|
|
What is Km dependent on? |
Only the substrate enzyme interaction, not the concentration of either |
|
|
What do you need to determine kcat that isn't needed for Km |
The enzyme concentration. |
|
|
When doing inhibition studies, what concentration of substrate should you use, and why? |
low levels of substrate ensure that the inhibitor is not out-competed |
|
|
When doing studies in microsomes or other biological environments, what should you be prepared to consider, and and how do you accomidate for it? |
Know that action by multiple enzymes is possible, to avoid this use specific inhibitors. |
|
|
What are the effects on Km and Vmax for competitive and non-competitive inhibitors |
Competitive inhibitors raise the Km (1+ [I]/[Ki]) non-competitive inhibitors lower the vmax (1 + [I]/Ki) |
|
|
What are the two metrics for inhibitor potency? |
I.C. 50 and KI I.C. 50 is definied as the concentration that reduces enzyme efficiency by 50% |
|
|
How does I.C. 50 relate to KI for competitive and non competitive inhibitors? |
for competitive inhibitors: I.C. 50 = Ki * ([I] + [S]/Km) for noncompetitive inhibitors: IC50 = Ki |
|
|
Do I.C. values depend on substrate concentration? |
for competitive inhibitors, but not for noncompetitive inhibitors |
|
|
What is the definition of a Mechanism Based Inhibitor (MBI) |
A MBI is a relatively unreactive compound that is converted by an enzyme into a species, which without prior release from the active site, binds covalently to the enzyme rendering it inactive. |
|
|
What are the 7 criteria for a compound to be determined a MBI? |
- Enyzme activity falls with time in MBI presence - Rate of inactivation is 1st order with time and a hyperbolic function of MBI for the active site of the enzyme -The inclusion of a normal substrate reduces the rate of inactivation via competition -Inactivation is irreversible -1:1 stoicheometry of enzyme and adduct -A catalytic step is required - Inactivation must occur prior to release |
|
|
What are the 3 types of adducts |
Heme pyrrole nitrogen alkylation protein alkylation MI complexes
|
|
|
How do you I.D. Heme pyroole nitrogen alkylation |
measure the CO complex of a heme in a competitive experiment. It will decrease with MBI treatment |
|
|
How do you I.D. protein adduction |
radiolabled MBI followed by a gel Mass spec / MS-MS |
|
|
How do you ID MI complexes? |
They are formed in time and can be measured with a UV/vis spec with difference spectroscopy (lambda = 456 nm) |
|
|
What is a partition ratio? |
Kp(products other than the inactive enzyme)/ Kinact(Dead enzyme) |
|
|
What type of MBI do epoxides normally take part in |
protein alkylation |
|
|
What is a MI complex |
It's a quasi irreversible inhibitor that binds to the heme iron. It produces a spectroscopically unique species. They can be regenerated if you reduce the iron. |
|
|
How do you characterize MBI's with kinetics? |
Multiple experiments where you very the amount of incubation and the amount of inhibitor |
|
|
what is lambda in terms of MBIs |
the rate of inactivation |
|
|
what is the relationship between lambda, Ki and K inactivation? |
lambda = K(degredation) = (Kinact * [I])/([I] + Ki) |
|
|
When do we care about MBI? |
when it causes much faster degredation of proteins than natural sources |
|
|
What is the general sequence of events for nuclear receptors (NR) that leads to increased transcription? |
-the NR begins associated with corepressors - inducer binds and NR dissociates -NR translocates to nucleaus _NR associates with dimerization partner -binding of heterodimer to response elements -release of corepressor proteins -Recruitment of coactivators and general transcription machinery |
|
|
What are some additional complexitires to nuclear receptor (NR) dynamics? |
-NR cross talk and ligand concentration dependent effects -NR expression variability -NR splice varients -tissue specific corepressors, coactivators, and transcription factors |
|
|
What causes the change in magnitude for responses between genes when activated by PXR |
the co-activator specific interactions (an indirect effect of the original ligand) |
|
|
Why do we care about induction |
it can change the amount of metabolism of a drug, thus affecting the amount of active species, and toxicity |
|
|
what determines the clinical significance of Induction |
the magnitude of change of an active species as a result of induction |
|
|
what is the definition of induction |
an increase in steady-state concentration of enzyme following exposure to an appropriate stimulus |
|
|
how does induction accelerate metabolism? |
an increase of Vmax |
|
|
what are the 2 main ways to cause induction |
increase of transcription or prevention of degredation |
|
|
why are some drugs that autoinduce their own metabolism so tricky? |
autoinduction causes a much longer time to reach steady state |
|
|
from a biochemical stantpoint, how do nuclear receptors increase transciption |
the binding of NR disrupts chromatin structure, facilitating the binding of the polymerase II complex and other transcription factors |
|
|
What NR regulate CYP 1A1/1A2/1B1 and what induces it |
AhR-ARNT, it is induced by antiestrogens and PAH |
|
|
What NR regulate 2B6/2C9/2C8 and what induces it? |
CAR-RXR(alpha), induced by androstanes, bile acids and phenobarbatal |
|
|
What NR regulates CYP4A and what compounds induce it? |
PPARalpha - RXR alpha Induced by fibrates ( fibrates are a class of amphipathic carboxylic acids. They are used for a range of metabolic disorders, mainly hypercholesterolemia, and are therefore hypolipidemic agents) and glitazones (a class of medications used in the treatment of diabetes mellitus type 2.) |
|
|
What does AhR stand for? |
aryl hydrocarbon receptor |
|
|
What physiological functions does CAR help regulate that encourages caution in inducing it? |
bilirubin clearance, bile acid detoxification, glucose control |
|
|
What structural feature of PXR leads to its promiscuity towards xenobiotics |
it has a large binding pocket |
|
|
what happens when an inducer is also an inhibitor of the enzyme it induces |
inhibition happens first, then induction. Overall the dynamics are much more complex |
|
|
Why is 3A4 more induceable than 3A5? Also, what NR induces it |
3A4 has proximal and distal response elements where 3A5 only has the proximal.
|
|
|
how are NR similar and different across species |
the DNA binding domain is fairly conserved, but the ligand binding domains are vastly different |
|
|
PXR activates multiple genes, but they have different responses, why? |
Binding different ligands and different heterodimer partners causes different affinities towards different genes |
|
|
What causes 2 different degredation half lives |
there are 2 degredation pathways: Ubiquitin and lysosome dependent |
|
|
How is CYP2E1 commonly induced |
Limitation of degredation (ethanol will bind and stabilize the protein, preventing lysozyme degredation) |

