![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
71 Cards in this Set
- Front
- Back
|
Major function of lipids: |
1. Structural component of membranes (phospholipids)
2. store metabolic energy and pr ovide padding (triacylglycerols) 3. Regulate metabolic activities (steroids) 4. Local hormones (eicosanoids) |
|
|
Structural Proteins
|
Made from long polymers |
|
|
Residue
|
an amino acid in a polypeptide chain
|
|
|
Glycoproteins
|
Proteins with carbohydrate groups attached
Components of cellular plasma membranes |
|
|
Proteoglycans
|
Mixture of proteins and carbohydrates
Consist of more than 50% carbohydrates Major component of extracellular matrix |
|
|
Cytochromes
|
Proteins which require a prosthetic (nonproteinaceous) heme group in order to function
Add color to cell Examples: Hemoglobin Cytochromes of electron transport chain in inner-membrane of mitochondria |
|
|
Conjugated Proteins
|
Proteins containing other nonproteinaceous components
|
|
|
Carbohydrates
|
Sugars or saccharides (-ose)
Empirical formula = C(H2O) Are ketones/aldehydes, water can be break the glycoside linkages of carbohydrates |
|
|
Anomeric Carbon
|
Cyclization of a carbohydrate causes the formation of 2 new diasteriomers. They differ in the position of the attachment of a certain group to the new stereocenter. The new stereocenter is referred to as the
anomeric carbon. To find it, locate lone oxygen on ring, then find alpha carbon with OH (anomeric carbon), not CH2-OH. |
|
|
Glucose
|
6-C carbohydrate
Account for 80% of carbohydrates absorbed by humans All digested carbohydrates reach cells as glucose Forms ring over chain form in aqueous solutions |
|
|
The 2 glucose anomers:
|
Ring form
Alpha-glucose: hydroxyl group on anomeric C (C #1) and methoxy group (C #6) are on opposite sides of ring Beta-glucose: hydroxyl group and methoxy group are on same side of ring |
|
|
Glycogen
|
alpha 1,6 (branch) and alpha 1,4 linkages
If sufficient ATP, glucose is polymerized to a polysaccharide (glycogen) or converted to fat Found in all animal cells, large amounts in liver and muscle cells |
|
|
Starch
|
2 forms:
1. Amylose (isomer of cellulose, alpha 1,4 linkages) 2. Amylopectin (resembles glycogen, different branching structure) Plants form starch from glucose |
|
|
Cellulose
|
Beta 1,4 linkages
Plants form cellulose from glucose Animals do not have enzymes to digest beta-linkages (bacteria inside animals do) |
|
|
3 components of Nucleotides
|
1. 5-C sugar (pentose)
2. Nitrogenous base 3. Phosphate group |
|
|
Nucleotides form polymers to create:
|
1. Nucleic acids
2. DNA (double stranded) 3. RNA (single stranded) |
|
|
Phosphodiester bonds
|
strong covalent bonds between a phosphate group and two 5-carbon ring carbohydrates (pentoses) over two ester bonds.
In DNA and RNA, the phosphodiester bond is the linkage between the 3' carbon atom of one sugar molecule and the 5' carbon atom of another, deoxyribose in DNA and ribose in RNA. |
|
|
Cyclic AMP
|
Nucleotide: Component in many second messenger systems
|
|
|
NADH & FADH2
|
Nucleotides: Coenzymes involved in Krebs cycle
|
|
|
Minerals
|
Dissolved inorganic ions inside and outside cell
By creating electrochemical gradients across membranes, they assist in transport of substances entering and exiting cells Can combine and solidify to give strength to a matrix (hydroxyapatite in bone) Act as cofactors assisting enzyme or protein function (iron in heme, prosthetic group of cytochromes) |
|
|
Enzymes
|
Globular proteins
Act as a catalyst = lowering activation energy for biological reactions and increasing reaction rates Not consumed or permanently altered by reactions Do not alter equilibrium of reaction |
|
|
Substrate
|
Reactant upon which an enzyme works
|
|
|
Active site
|
Position on enzyme to which subtrate binds with numerous covalent bonds
|
|
|
Lock & Key Theory
|
Example of enzyme specificity
Active site of enzyme has specific shape (lock) that only fits a specific substrate (key) Model does not explain all enzymes |
|
|
Induced Fit Model
|
Shape of both enzyme and substrate are altered upon binding
Alteration helps reaction proceed and increases specificity |
|
|
Saturation Kinetics
|
As relative concentration of substrate increases, rate of reaction also increases, but to a lesser degree until a maximum rate (Vmax) is achieved
More substrate is added and therefore must wait in line for free enzyme Vmax is proportional to substrate concentration |
|
|
What affects enzyme reactions?
|
1. Substrate concentration
2. pH (optimal pH) 3. Temperature (as temperature increases, reaction rate increases until enzyme denatures) |
|
|
Cofactor
|
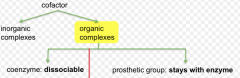
Non-protein component that helps enzymes reach their optimal activity
Can be coenzymes or metal ions |
|
|
Coenzymes
|
2 groups:
1. Cosubstrates 2. Prosthetic groups Organic molecules |
|
|
Cosubstrates
|
Category of cofactor
Reversibly bind to a specific enzyme and transfer some chemical group to another substrate Reverted to original form by another enzymatic reaction (distinguishing it from a normal substrate) Example: ATP |
|
|
Prosthetic groups
|
Remain covalently bound to enzyme throughout reaction
Emerge from reaction unchanged Example: Heme & metal ions (can act alone or with prosthetic group) |
|
|
Vitamins
|
Can be coenzymes
Are essential organic molecules: cannot be produced by body |
|
|
Apoenzyme
|
Enzyme without cofactor
Completely nonfunctional |
|
|
Holoenzyme
|
Enzyme with cofactor
|
|
|
3 mechanisms of enzyme inhibition
|
1. Irreversible inhibition
2. Competitive inhibition 3. Noncompetitive inhibition |
|
|
Irreversible Inhibitors
|
Agents which bind covalently to enzymes and disrupt their function
Highly toxic (penicillin) |
|
|
Competitive Inhibitors
|
Compete with substrate by binding reversibly with noncovalent bonds to active site
Can overcome inhibition by increasing concentration of substrate Resemble substrate |
|
|
Noncompetitive Inhibitors
|
Bind noncovalently to enzyme at spot other than active site and change conformation of enzyme
Do not prevent substrate from binding Do not resemble substrate Act on more than one enzyme Cannot be overcome by excess substrate Do not lower enzyme affinity for substrate |
|
|
4 mechanisms by which enzymes are regulated:
|
1. Proteolytic cleavage (irreversible covalent modification)
2. Reversible covalent modification (AMP, phosphorylation, or hydrolysis) 3. Control proteins (activate or inhibit activity) 4. Allosteric interactions (modification of enzyme) |
|
|
Zymogen (proenzyme)
|
Inactive form of enzyme
Specific cleavage of peptide bonds irreversibly activates enzyme Activation may be instigated by other enzymes Example: pepsinogen (pepsin) |
|
|
Allosteric Interactions
|
Modification of enzyme conformation resulting from binding of activator or inhibitor at specific binding site on enzyme
Can be allosteric activators or allosteric inhibitors of an enzyme, competitive or noncompetitive |
|
|
Negative feedback (feedback inhibition)
|
Product of downstream reaction inhibits enzymatic activity in upstream reaction
Provides shut-down mechanism for series of enzymatic reactions when it has produced sufficient amount of product Do not resemble substrate of enzymes that they inhibit - bind to enzyme and cause conformational change (allosteric regulation) |
|
|
Positive feedback
|
Product activates enzyme
|
|
|
Positive cooperativity
|
1st substrate changes shape of enzyme allowing other substrates to bind more easily
|
|
|
Negative cooperativity
|
1st substrate changes shape of enzyme, inhibiting substrate binding
|
|
|
Enzyme Nomenclature
|
Suffix "ase" is added to end of substrate upon which enzyme acts
(acetylcholinesterase acts upon the ester group in acetylcholine) |
|
|
Lyase (an enzyme classification)
|
(aka synthase) Catalyzes addition of 1 substrate to a double bond of a 2nd substrate
Example: ATP synthase |
|
|
Ligase (an enzyme classification)
|
(synthetase) Governs an addition reaction
Requires energy from ATP or other nucleotide |
|
|
Kinase
|
Enzyme which phosphorylates something
|
|
|
Phosphatase
|
Enzyme which dephosphorylates something
|
|
|
Products and reactants for respiration? What kind of reaction is it?
|
Glucose + O2 -> CO2 + H2O
Combustion reaction |
|
|
Aerobic respiration
|
Oxygen is used
Produces 32-36 ATP/glucose |
|
|
Glycolysis equation
|
Glucose (6C) + 2NAD+ + 2 ADP + 2 Pi -> 2 pyruvate (3C) + 2 ATP + 2 NADH + 2 H2O
Operates in both presence and absence of oxygen, occurs in cytosol |
|
|
Substrate level phosphorylation
|
Formation of ATP from ADP and Pi using the energy released from the decay of high energy phosphorylated compounds
No use of energy from diffusion |
|
|
Fermentation
|
Anaerobic respiration
1. Glycolysis (glucose -> 2 pyruvate) 2. Reduction of pyruvate to ethanol or lactic acid (CO2 expelled as waste product) 3. Oxidation of NADH back to NAD+ Yeast and microorganisms produce ethanol, humans produce lactic acid Takes place when: 1. Organism is unable to assimilate energy from NADH and pyruvate 2. Organism has no oxygen available |
|
|
Mitochondria membrane permeability
|
Outer membrane: permeable to pyruvate and NADH (facilitated diffusion)
Inner membrane: permeable to pyruvate (fac. diff.) but NADH may require hydrolysis of ATP |
|
|
Once inside matrix, pyruvate is...
|
converted to acetyl CoA (2C) in a reaction that produces NADH & CO2
CoenzymeA transfers 2-C from pyruvate to 4-C oxaloacetic acid to make citrate, Krebs cycle |
|
|
Krebs Cycle (citric acid cycle)
|
Each turn produces:
1. 1 ATP 2. 3 NADH 3. 1 FADH2 4. 2C released as CO2 5. Oxaloacetic acid regenerted to begin cycle again Process of ATP production is substrate-level phosphorylation 1x glucose = 2x turns of Krebs cycle |
|
|
What is the Electron Transport Chain (ETC)?
|
A series of proteins, including cytochromes with heme, in inner-membrane of mitochondria
1st protein complex oxidizes NADH by accepting its high energy electrons Electrons are passed down protein series and ultimately accepted by oxygen to form water As electrons are passed along, protons are pumped into intermembrane space for each NADH. Establishes proton gradient which propels protons through ATP synthase to manufacture ATP (oxidative phosphorylation) 2-3x ATP made from each NADH 2x ATP made from FADH2 (reduces protein) |
|
|
Proton-motive force
|
Proton gradient established from pumping out of protons into intermembrane space, which propels protons through ATP synthase to make ATP (oxidative phosphorylation!)
|
|
|
ATP synthase
|
Makes ATP by oxidative phosphorylation
|
|
|
Acidic Alpha AAs?
|
AG
Aspartic acid Glutamic acid |
|
|
basic alpha aa's?
|
HAL
Histidine Arginine Lysine |
|
|
Nonpolar alpha aa's?
|
VIPMALT PG
Valine Isoleucine Proline Methionine Alanine Leucine Tryptophan Phenylalanine Glycine |
|
|
Polar alpha aa's?
|
STAT GC
Serine Threonine Asparagine Tyrosine Glutamine Cysteine |
|
|
Cysteine
|
Has SH (sulfur) group, forms disulfide bonds
|
|
|
Proline
|
Creates a kink, never involved in secondary structure
|
|
|
Primary Structure of Protein
|
Determines ultimate configuration of protein
held together by peptide bonds primary structure is rarely affected by denaturing agents |
|
|
Secondary structure
|
Alpha helix, beta sheets
Held together by H-bonds between different AA's |
|
|
Tertiary structure
|
Based on disulfide bonds, hydrophobic/philic effects, ionic bonding (between acidic and basic side chains), hydrogen bonding, and van der waals
|
|
|
Quaternary structure
|
Consists of multi-subunit proteins
Bonds aren't internal: bonds are made between other proteins |

