![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
20 Cards in this Set
- Front
- Back
|
What is the etiological agent in AIDS
|
the retrovirus HIV-1
|
|
|
How are retroviruses characterized?
|
Retroviruses contain an RNA genome and an RNA dependent DNA polymerase (reverse transcriptase)
|
|
|
Why is HIV unique
|
It targets the immune system, forms significant amounts of progeny in vivo, and is sexually transmitted.
|
|
|
What is the structure of HIV?
|
It is a particle surrounded by a lipid bilayer derived from the host cell membrane
|
|
|
How do we calculate the length of fragments based on their migration?
|
Fragments migrate inversely proportional to the log10 of the molecular weight. So find the migration equation for that gel, then take the inverse log10
|
|
|
Be able to differentiate linear, supercoiled, and nicked circular DNA in an agarose gel
|

|
|
|
What factors effect the migration of DNA in an agarose gel?
|
Agarose Concentration (one percentage does not fit all bands)
Voltage (increased voltage leads to decreased resolution) Fragment length |
|
|
What are restriction enzymes?
|
Enzymes that cut phosphodiester bonds in the backbone of DNA molecules at specific recognition nucleotide sequences. Require metal ions as cofactors (Mg²⁺).
|
|
|
How are restriction enzymes named?
|
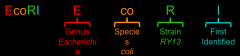
|
|
|
What are the four different types of restriction enzymes?
|
Type I: cleaves at sites remote from recognition site, require ATP for enzymatic activity
Type II: cleave within a short specific recognition sequence (4 - 8bp), require Mg2+ Type III: cleaves at short distance from recognition site, require ATP for enzymatic activity Type IV: have two enzymatic activities (methyl transferase and endonuclease), do not require ATP for enzymatic activity |
|
|
What is plasmid DNA?
|
Circular molecule
Independent replication from chromosomal DNA 1 to over 500 Kbp Conformations: Nicked, Relaxed, Supercoiled |
|
|
Lambda DNA
|
From a phage Lambda virus
48,000bp Linear Usually used as a standard weight marker for DNA |
|
|
What is a mole?
|
1 Mole is the number of atoms in a mass of 12 g of isotope Carbon-12 (C12) = 6.022 x 1023 atoms = Avogadro’s number
•The mass of one mole of a substance, expressed in grams, is exactly equal to the substance's mean molecular weight. |
|
|
What is molecular weight?
|
The molecular weight (MW, or gram molecular weight) of a substance is equivalent to the sum of its atomic weights.
|
|
|
What is Normality?
|
Normality is always a whole-number multiple of molarity. Normality = equiv # x Molarity
|
|
|
What are synthetic vectors used for?
|
Synthetic vectors designed for specific functions:
Cloning sequences. Protein expression. RNAi. |
|
|
Name the steps for isolating plasmid DNA:
|
1. Harvest cells by centrifugation.
2. Remove medium 3. Resuspend cells in buffer. 4. Add lysis buffer (alkaline lysis). 5. Add neutralization buffer. 6. Mix to precipitate debris. 7. Centrifuge to clear solution (plasmid DNA). 8. Transfer supernatant to fresh tube. 9. Precipitate DNA with isopropanol (DNA visible?). 10. Centrifuge to collect DNA. 11. Drain tube and wash pellet with ethanol. 12. Centrifuge, drain and dry pellet. 13. Resuspend DNA in TE. 14. Prepare gel and run test aliquot. |
|
|
Name the steps in isolating chromosomal DNA:
|
1. Add EDTA Buffer to tube of cells.
2. Add RNase solution to tube. 3. Add Sarkosyl solution (detergent). 4. Add Protease solution. 5. Incubate to digest cell wall, RNA and proteins. 6. Add NaCl solution. 7. Transfer solution to flask. 8. Carefully overlay isopropanol solution onto salt solution. 9. Swirl glass rod to “spool” DNA onto rod. 10. Transfer rod to tube of Tris buffer. 11. Twirl rod to dislodge DNA |
|
|
Where does Taq P. originate from?
|
Taq Polymerase is from hot spring bacteria (Brock and Hudson) – does not denature at 94°C.
|
|
|
What is VNTR?
|
VNTR: Variable Number of Tandem Repeats
the variability or differences in VNTR alleles are in the "number of tandem repeats." |

