![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
72 Cards in this Set
- Front
- Back
|
Adenine
|
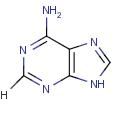
Name
|
|
|
Cytosine
|
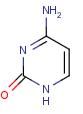
Name
|
|
|
Guanine
|
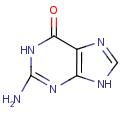
Name
|
|
|
Thymine
|
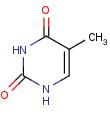
Name
|
|
|
Uracil
|
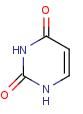
Name
|
|
|
2'-deoxyribose
|
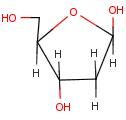
Name
|
|
|
Purine linkage (1-9 linkage)
|
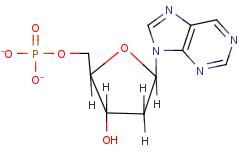
Name linkage
|
|
|
Pyrimidine Linkage (1-1 Linkage)
|
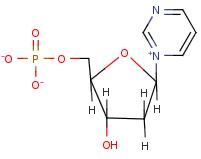
Name linkage
|
|
|
Ribose
|
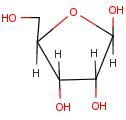
Name
|
|
|
Nucleotide
|
Unit structure of DNA. Composed of nitrogen base, sugar, and phosphoric acid
|
|
|
Deoxyribonucleic Acid
|
Chemical unit of heritable information
|
|
|
Erwin Chargaff
|
Described fundamental ratios of nitrogenous bases in DNA
|
|
|
Tetranucleotide Hypothesis
|
4 bases are present in equal amounts. Tested by Chargaff
|
|
|
Chargaff's Rules
|
#A=#T, #G=#C
(A+G)=(T+C) %(G+C)≠%(A+T) |
|
|
Rosalind Franklin
|
Used x-ray diffraction to obtain pictures of DNA molecule. Recognized DNA as helical
|
|
|
Periodicity of DNA
|
Bases are 3.4Å apart with 10 bases per turn.
|
|
|
Model of DNA
|
- Right handed helix
- 1 complete turn= 34Å - Diameter= 20Å - Space between pairs= 3.4Å - 10 bases per turn - Antiparallel - Major and minor grooves alternate |
|
|
Avery, Macleod, and McCarty
|
-Made the TRANSFORMING PROPERTY- heritable properties are carried on DNA
-Used mice to show that DNA is capable of transformation |
|
|
Hershey and Chase
|
-DNA is the bio-molecule of heredity
-Used bacteriophage to show that DNA is genetic material -Protein is not the genetic material |
|
|
Erwin Chargaff
|
Base composition/ chemistry of DNA
|
|
|
Watson and Crick
|
Chemical components, physical structure, and molecular form of DNA
|
|
|
Classic form of DNA
|
Beta form
|
|
|
DNA Sequencing
|
The determination of the precise sequence of nucleotides in a sample of DNA
|
|
|
Common method of DNA sequencing
|
Chain termination
|
|
|
Chain termination occurs with
|
dideoxynucleotides
|
|
|
Needed for DNA Sequencing
|
1) ssDNA template
2) Primer 3) DNA polymerase 4) dNTP's 5) Appropriate dideoxynucleotides |
|
|
Electrophoresis
|
Uses electric field to seperate fragments based on size and length
|
|
|
AZT
|
Slows down replication
|
|
|
Alpha form of DNA
|
- High slat, dehydrated conditions
- More compact- 11 bp/turn - 23Å diameter |
|
|
Z form of DNA
|
- Zig zag conformaion
- No major grooves - Left handed helix |
|
|
Function of DNA
|
Genotype- form of genetic info
|
|
|
Function of RNA
|
Intermediate of phenotype, sometimes genotype
|
|
|
Bases of DNA
|
A=T, C=G
|
|
|
Bases of RNA
|
A=U, C=G
|
|
|
Sugar of DNA
|
2' deoxyribose
|
|
|
Sugar of RNA
|
Ribose
|
|
|
Unit structure of DNA
|
Nucleotides
|
|
|
Unit structure of RNA
|
Nucleotides
|
|
|
Gross structure of DNA
|
Always double stranded helical
|
|
|
Gross structure of RNA
|
Generally single stranded linear, when genotype, can be double sranded helical
|
|
|
Genome
|
Total number of nucleotides present in an organism that constitutes genetic information; diploid content of genetic information
|
|
|
Genome Complexity
|
Organizaion of nucleotide; biological properties
|
|
|
Genome Size
|
Number of nucleotides
|
|
|
Reassosciation Kinetics
|
Isolate DNA -> Fragment into small peices -> Heat/Denature -> Cool/Renature -> Fragments reassociate by complimentarity
|
|
|
Reassociation is dependent on
|
Genome size and complexity
|
|
|
Fast Reassosciation
|
Short repetitive sequences
|
|
|
Slow Reassociation
|
Longer lengths of DNA with increased complexity
|
|
|
Cot curve shape
|
Genome Complexity
|
|
|
Cot curve length
|
Genome size
|
|
|
Highly Repetitive DNA
|
- Non-coding
- Rapid reassociation - 5-10 bp in length - 5-10% of genome - Around centromeres and telomeres - Maintains chromosome morphology |
|
|
Moderately Repetitive DNA
|
- Non-coding
- 150-500 bp in length - 5-10% of genome - Repeated 700-900,000 times - Non coding- mutation buffers |
|
|
Unique DNA
|
- Coding- genotype
- 1,000-15,000 bp in length - 35,000 genes in humans - 1-5% of total nucleotides |
|
|
C-value
|
The amount of DNA contained in the haploid genome of a species
|
|
|
C-value Paradox
|
Excess DNA is present that does not seem to be essential to the development or evolutionary divergence of eukaryotes
|
|
|
G2 Phase
|
Cells prepare for division
|
|
|
G1 Phase
|
Cells sit- DNA is stringy
|
|
|
S/R Phase
|
- DNA makes copies and sister chromatids
- DNA synthesis and replication |
|
|
M Phase
|
Mitosis
|
|
|
Semi-conservative Replication
|
Watson and Crick proposed form. Following synthesis of the new strands, each parent strand links with a new strand in complimentary fashion
|
|
|
Conservative Replication
|
Synthesis follows Watson and Crick's model however by some mechanism, each parental strand reforms together while the 2 new strands also link together
|
|
|
Dispersive Replication
|
Each parental strand goes through a cleavage process followed by reforming of all strands together
|
|
|
Replicated Homologs
|
Sister chromatids
|
|
|
Replicon
|
Site of replication
|
|
|
Arthur Kornberg
|
DNA is universal/ highly conserved
|
|
|
Replication always occurs in which direction?
|
5' -> 3'
|
|
|
DNA Gyrase
|
- Member of the family of topoisomerases
- Reduces super-cooling of DNA |
|
|
Helicase
|
Opens DNA strands making them ssDNA
|
|
|
Primase
|
Lays down an RNA primer
|
|
|
DNA Polymerase
|
Binds RNA primer at 3' OH and makes new strands
|
|
|
Ligase
|
Joins together the newly synthesized strands of DNA by sealing the phosphodiester bonds
|
|
|
Telomerase
|
Fills in any gaps created at tips of the 3' ends in the old DNA strand
|
|
|
DNA Replication
|
1) Helicase binds to DNA strands and pull them apart
2) Primase recognizes origin points, binds to ssDNA, lays down RNA primer 3) Synthesis in 5' -> 3' direction 4) DNA pol removes primers and replaces them with DNA 5) Ligase fills gaps between fragments 6) Telomerase binds at 3' end to remove unmatched pairs |

