![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
143 Cards in this Set
- Front
- Back
|
The table above gives the results from intercrossing beetles with seven forms of mutant elytra (striated, punctate, rugose, wrinkled, fuzzy, glabrous, shiny, rough, scapose). An + in the table indicates that the offspring had a wildtype phenotype, a - indicates non-wildtype elytra. On these results, the cross between glabrous and fuzzy is best explained as:
|
complementation
|
|
|
In a human population the genotype frequencies at on locus are f(aa) = 0.51, f(Aa) = 0.43, and f(AA) = 0.06. The frequency of the A allele is_____?
|
0.285
|
|
|
In Drosophila the sv/+ locus is normally located at the end of a chromosome arm. The recessive allele sv shortens and flattens the bristles. In a certain
sv/+ heterozygotes, the + allele is relocated next to the centromere by an inversion. This fly showed patches of shaven and wild type bristles on its body. This is an example of: |
position effect variegation
|
|
|
When an autotetraploid is backcrossed to its parent species, sterile offspring are produced. Such sterile offspring can be best represented by:
|
3n
|
|
|
In a particular strain of tomato, fruit color is determined by the following three alleles: R = red, r = albino, and rm = albino (R with Te inserted). A cross
is made between rm/r Te+/Te+ and R/r Te/Te+. What will be the phenotypic ratio in the progeny? [Assume that Te does not jump into the non-inserted R allele or break the chromosome containing the R allele.] |
1/2 red, 1/4 albino, 1/4 albino with red spots
|
|
|
The norm of reaction is important because:
|
the range of phenotypes produced by a genotype will vary depending on the environment
|
|
|
Suppose you have a large population of flour beetles. Red body color is due to a dominant allele, R; individuals who are genotypically RR or Rr are red.
Individual which are homozygous recessive rr have black body color. If the population is in Hardy-Weinberg equilibrium, with f(R) = 0.55 and f(r) = 0.45, what are the expected frequencies of the red and black phenotypes? |
80% red, 20% black
|
|
|
What is the name of the enzyme that, during DNA replication, extends the lagging strand from the 3' end of each RNA primer?
|
DNA polymerase III
|
|
|
What would happen during DNA replication in a cell lacking topoisomerase?
|
DNA ahead of the replication fork would become increasingly tightly coiled.
|
|
|
What observation(s) were crucial in allowing Watson and Crick to propose the double helix structure of DNA?
|
Chargraff's rules concerning the relative frequency of different nucleotides and Franklin's X Ray diffraction of the DNA molecule.
|
|
|
Which of the following distinguishes RNA from DNA?
|
RNA has an oxygen at the 2’ position of ribose.
|
|
|
In what way did the Avery, MacLeod and McCarty experiment build on Griffith’s experiment? They:
|
Identified DNA as the transforming factor through a process of elimination.
|
|
|
This parental helix of DNA is replicated in a test tube containing all the proteins necessary for DNA replication, GTP (guanosine triphosphate) and radioactive CTP (cytosine triphosphate - 32P):
5’-GGGGGGGGGGG-3’ 3’-CCCCCCCCCCC-5’ What pattern of radioactivity would you expect among the new helices? |
Only 1 of the new helices would be radioactive it is possible to predict which one
|
|
|
In E. coli, what do sigma factors do during transcription?
|
Help the holoenzyme to bind to the promoter
|
|
|
In prokaryotes, the functional equivalent of the TATA binding protein is:
|
Sigma factor.
|
|
|
You delete a gene coding one of the proteins required for splicing (removing) introns from pre-mRNA. This deletion mutation stops growth of a yeast cell. Why?
|
Failure to remove introns will lead to non-functional proteins because their primary sequence will be derived from introns as well as exons.
|
|
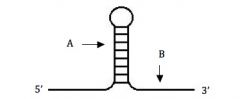
Which answer combines the correct responses to the questions below?
(i) What is the name of the structure shown below? (ii) What is its function in transcription? (iii) What is the probable base composition of the region marked by the “A” arrow? (iv) What is the probable base composition of the region marked by the “B” arrow? |
(i) Hairpin loop; (ii) Termination of transcription in prokaryotes; (iii) G/C rich; (iv) string of U’s.
|
|
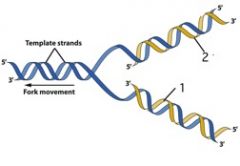
In this picture of DNA, which number identifies the leading strand and how can you tell?
|
1 because the 3’ end is extended in the same direction as the replication fork
|
|
|
What is the name of the sequence region recognized by the bacterial ribosome?
A. -10 and -35 consensus sequences. |
Shine-Dalgarno
|
|
|
Which of the following constitutes the primary structure of a protein?
|
The linear sequence of amino acids in a polypeptide chain.
|
|
|
What experimental observation addressed the likelihood of the genetic code being overlapping?
|
A mutation changing a single nucleotide changed one amino acid in the corresponding protein.
|
|
|
Which of the following is an important feature of the genetic code?
|
-It is a triplet code.
-Each codon specifies no more than one amino acid. -It is degenerate. -It is non-overlapping. |
|
|
What answer represents the most likely order of the severity of the phenotypic effect resulting from the different types of mutations listed below. (Answers are ranked from most to least severe.)
i. Insertion of a nucleotide causing a shift in the codon reading frame. ii. Nucleotide substitution that does change the amino acid a codon codes for. iii. Nucleotide substitution that does not change the amino acid a codon codes for. |
i>ii>iii
|
|
|
A newly discovered set of genes – the hou operon – are transcribed from the same promoter. They allow the bacteria to use a rare sugar, houstose. This sugar doesn’t give much energy when it is broken down, but, when nothing else is available, it is better than nothing. In what combination of environmental conditions would you expect bacteria to express the hou operon? (Assume that conserving energy used in expressing a gene is important – if a bacteria does not need to express a gene, it won’t.)
|
In the presence of houstose and the absence of any other more effective nutrient source.
|
|
|
The lac repressor (LacI) binds to:
|
Lactose and DNA.
|
|
|
Does the lac repressor (LacI) act as a cis or a trans regulator?
|
Trans.
|
|
|
A mutant strain of E. coli has the following genotype at its lac operon (assume all other genes are functional): I+ P+ O- Z+ Y+ A+ (I = repressor; P = promoter; O = operator; Z,Y,A = structural genes). Which of the following answers best match the environments in which the operon will be expressed at a high level?
|
Glucose is absent, regardless of the presence of lactose.
|
|
|
What is the best explanation for both positive (cAMP-CAP) and negative (LacI) regulation being needed to ensure that the lac operon is only expressed when it is needed?
|
Each control system (cAMP-CAP and LacI) is sensitive to only one environmental condition (glucose or lactose) – both controls are needed for regulation by both glucose and lactose.
|
|
|
How does tryptophan inhibit transcription of the trp operon through attenuation?
|
The ribosome NOT pausing at two consecutive trp codons in the leader region causes mRNA to fold in a way that terminates transcription by RNA polymerase
|
|
|
In yeast, the Gal structural genes are controlled by the regulatory proteins Gal4, Gal80 and Gal3. Gal4 is a positive regulator which binds to activation sequences upstream of the Gal structural genes. Gal80 binds to Gal4 and inhibits the ability of Gal4 to activate gene expression. In combination with galactose, Gal3 reduces Gal80 inhibition and allows Gal4 to activate Gal structural gene expression. How would you expect the expression of Gal structural genes to depend on galactose in a Gal80- mutant?
|
Expression of Gal structural genes would be ON, regardless of the presence of galactose.
|
|
|
What are the base (default) positions of bacterial and eukaryotic promoters – that is, will gene transcription be ON or OFF in the presence of RNA polymerase and any accessory proteins but in the absence of any specific transcription factors?
|
Bacterial promoters are ON and Eukaryotic promoters are OFF by default
|
|
|
The hunchback gene has an enhancer containing 3 binding sites for the transcription factor Bicoid. How could the loss of a single DNA binding site – leaving 2 others intact – act to reduce gene expression of hunchback by more than 1/3?
|
Bound Bicoid transcription factor molecules interact cooperatively to cause maximal gene expression.
|
|
|
Which of the below are types of analyses that you could perform if you knew the entire sequence of a genome?
|
All of the above
|
|
|
Which of the following is a key advantage of reverse genetics in comparison with forward genetics? (Reverse genetics: observing the phenotypic consequence of disruption of a specific gene. Forward genetics: Identifying the genetic changes that are associated with a particular phenotype.)
|
Only reverse genetics can establish cause and effect between a genetic change and a phenotypic consequence.
|
|
|
Central Dogma
|
DNA is information coding molecule
-it gets replicated (DNA->DNA) -passes on information to intermediary (DNA->RNA) *transcription Translation- information is conversion of the RNA information to Protiens |
|
|
Bacterial Transcription
|
2 dna strands separate (replication bubble) - one strand acts as template.
RNA polymerase assembles an RNA strand using DNA template strand. RNA (U, A, C, G vs. A, T, G, C for DNA) |
|
|
RNA Replication assembly proceeds at
|
5' to 3' (new nucleotides to 3' end)
|
|
|
In DNA replication the DNA strand that is not used as a template is called the _____ __________.
|
coding strand
|
|
|
Classes of RNA
|
INFORMATIONAL:
Messenger RNA FUNCTIONAL: transfer (tRNA) ribosomal (rRNA) small nuclear (snRNA) micro (miRNA) small interfering (siRNA) |
|
|
Informational RNA
|
intermediate between DNA sequence and protien sequence
|
|
|
Functional RNA
|
carry out some physiological role as RNA molecules
|
|
|
Central Dogma: Condensed
|
DNA Polymerase - REPLICATION
RNA Polymerase - TRANSCRIPTION Ribosome - TRANSLATION |
|
|
DNA:A
RNA:__ |
U
|
|
|
DNA:T
RNA:__ |
A
|
|
|
DNA:G
RNA:__ |
C
|
|
|
DNA:C
RNA:__ |
G
|
|
|
__________RNA polymerases can __________ transcribe a _______ gene.
|
Multiple, simultaneously, single
|
|
|
Three stages of Transcription
|
-initiation
-elongation -termination |
|
|
What suggested that RNA might serve a role as an intermediate between DNA and proteins?
|
Scientists noticed a burst of RNA production immediately following bacteriophage infection.
|
|
|
RNA is usually _______ stranded, but still able to base pair (with itself) through _________ _________, this usually happens '____-__________' which give RNA _____________ ________.
|
single stranded (ssRNA), hydrogen bonds, 'intra-molecularly', specific shapes
|
|
|
DNA has ______sugar and RNA has _______ sugar in each nucleotide
|
deoxyribose, ribose
|
|
|
RNA can do _______ _______ ______(as can protien) and DNA cannot.
|
catalyze chemical reactions
|
|
|
ribozyme
|
RNA that catalyzes reactions
|
|
|
U=______ -> for RNA
T=______ -> for DNA |
Uracil
Thymine |
|
|
DNA structure frame used to organize the flow of information transfer inside a cell was proposed by:
|
Crick
|
|
|
Step 1 of transcription:
DNA-DNA |
DNA strands locally separate (a bubble) - one strand acts as template the other is coding strand
|
|
|
Step 2 of transcription: RNA
DNA-RNA |
An RNA strand is assembled using the template DNA strand as a guide. 5'-3' of the DNA (new nucleotides added to 3' end of RNA growing chain)
-assembly done by RNA polymerase |
|
|
Transcription: Initiation
(prokaryote transcription initiation) |
-promoter indicates, to RNA polymerase, where to begin RNA synthesis (promoters located upstream of the region of DNA to be transcribed
|
|
|
Consensus
|
average sequences that seems to be recognized by RNA polymerase - the sequences are the core of the promoter.
-TATAAT (-15) -TTGACAT (-35) Usually promoters with sequence close to the consensus will be better recognized by RNA polymerase and transcribed at a higher rate. |
|
|
Initiation:
|
-RNA polymerase complex scans DNA and recognizes a promoter (specifically the -10 and -35 sequence stretches)
-DNA is unwound in this location to reveal single stranded DNA (ssDNA) -Transcription begins from the +1 position - located just downstream of the promoter |
|
|
5' untranslated region
|
(5' UTR) - upstream region of mRNA between transcription start and start of gene is called this. This has sequence important for translation that the ribosome recognizes.
|
|
|
RNA polymerase holoenzyme
|
multi-protein enzyme that consists of:
-2 alpha units, 1 beta unit, 1 beta' unit, 1 omega unit (core enzyme - nessasary for function) -sigma factor: directs enzyme to promoter |
|
|
Sigma factor 70
|
directs RNA polymerase to the promoter ( -10 and -35 upstream region before the start of transcription)
-sigma factor is released once the transcription begins |
|
|
RNA must _______ the _____ _______ as it travels down the gene being transcribed - forming a '_________ ______'
|
unwind, DNA helix, 'transcription bubble'
|
|
|
Single free RNA nucleotides are synthesized in other reactions in the cell. What happens when they com into contact with the RNA polymerase
|
When complimentary match to the ss DNA occurs the RNA polymerase adds it to the 3' end of the RNA chain
|
|
|
Signals that cause RNA polymerase to release from the DNA helix:
|
Intrinsic and Rho-dependant
|
|
|
Intrinsic
|
strong hairpin/ stem-loop (intra molecular bonding) RNA pairs with itself. structure followed by a region of weak binding between RNA and DNA destablizies the RNA polymerase and it is released from the DNA helix
|
|
|
Rho-dependant:
|
rho protein binds to the RNA strand and travels down until it reaches the RNA polymerase and displaces it from the DNA helix
|
|
|
which has more genes? bacteria or eukaryotes?
|
eukaryotes (1-10 times as many)
|
|
|
Three RNA polymerase
|
RNA Polymerase I
RNA Polymerase II RNA Polymerase III |
|
|
RNA polymerase I
|
transcribes ribosomal genes (rRNA)
|
|
|
RNA Polymerase II
|
transcribes protein coding genes
|
|
|
RNA Polymerase III
|
transcribes small functional RNA genes [e.g. transfer RNA genes - (tRNA)]
|
|
|
In eukaryotes transcription occurs where?
|
in the nucleus, where the DNA is
|
|
|
Because transcription for eukaryotes occurs in the nucleus, how does this affect RNA transcripts.
|
most RNA transcripts need to be exported outside the nucleus to the cytoplasm where translation takes place
|
|
|
What must happen to RNA transcripts before exportation outside of the nucleus to the cytoplasm
|
the transcripts must be processed
|
|
|
pre-mRNA
|
RNA before processing the transcript - in the nucleus
|
|
|
mRNA
|
RNA after transcript is processed-exported to the cytoplasm
|
|
|
general transcription factors
|
eukaryotes use to recognize promoter regions (similar to sigma factor in prokaryotes) protein that first identifies the promoter "TATA binding protein"
|
|
|
GTF names are
|
TFIIA, and TFIIB (transcription factor of RNA polymerase)
|
|
|
Eukaryote transcription elongation and processing
|
Capping, Cleavage and polyadenylation
|
|
|
Capping
|
5' -cap: cap is added to prevent degradation
|
|
|
Cleavage and polydenylation
|
3' poly(A) tail: when RNA polymerase II reaches a specific termination sequence it stops and a 150-200 adenine nucleotides are added
|
|
|
Splicing
|
section of the RNA transcript that are not meant to code for protein (introns) are removed to create a continuous stretch of coding RNA (exons)
|
|
|
What does splicing cause in reference to the pre-mRNA?
|
Protien predicted from a pre-mRNA differs from the protein that is actually observed.
|
|
|
In different tissues can Alternative Splices be made?
|
Yes, creating as much as 100 different proteins (an enormous variety from one gene)
|
|
|
What identifies introns as regions of pre-mRNA to be removed?
|
by comparing pre-mRNA with mRNA. regions ALWAYS missing are introns.
|
|
|
snRNPs
|
recruit additional proteins to form a complex called spliceosome
|
|
|
spliceosome
|
removes the intron and joins the exons together.
|
|
|
siRNA
|
prevent translation of mRNA- preventing expression of the corresponding protein
|
|
|
DICER
|
enzyme that produces siRNA in short 20-25 bp fragments
|
|
|
RISC
|
binds to ssRNA fragments and degrades mRNA that they pair with (complementary)
|
|
|
What is the general structure of protein (amino acid polymers)?
|
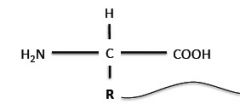
R group can be any of 20 that distinguish the different protein amino acids
|
|
|
what are proteins
|
proteins are polymers of amino acids
|
|
|
How are amino acids formed
|
through peptide bonds linking amino groups (NH2) of one group to the Carboxyl group (COOH) of another
|
|
|
chain of amino acids called?
|
polypeptide - called protein when complete
|
|
|
What is the major type of functional molecule in cells?
|
proteins
|
|
|
what do proteins need in order to be functional?
|
they need to be able to fold into a particular structure.
|
|
|
Four Level of structures for proteins
|
primary, secondary, tertiary, quaternary
|
|
|
primary structure
|
sequence of amino acids
|
|
|
secondary structure
|
particular shapes that local regions of a protein fold into (alpha helix and beta sheets)
|
|
|
tertiary
|
overall shape of protein
|
|
|
quaternary
|
multipe tertiary proteins (subunits) come together to make a functional protein
|
|
|
What determines structure
|
sequence
|
|
|
what determines function
|
structure
|
|
|
One gene - one protein
|
Beadle and Tatum - proposed each gene encodes (somehow) one protein
|
|
|
Yanofsky-1963
|
"changes in the gene were about the same position as the changes in the protein"
-Indicated that DNA and protein sequences were CO-LINEAR. |
|
|
overlapping code prediction is that a ______ mutation will change _______ amino acids.
|
single, multiple
|
|
|
observation of overlapping code is?
|
single mutations change a single amino acid and therefore the code is non-overlapping
|
|
|
Genetic code is overlapping or nonoverlapping?
|
non overlapping
|
|
|
How can a sequence of 4 letters (DNA bases) be translated into one with 20 letters (amino acids)
|
code length is 1 therefore the amino acids that can be coded for are 4 to the power of 1 = 4 and so forth. Therefore 4 to the power of 3= 64 so need a codon at least 3 nucleotides (bases) long to code all amino acids ( a triplet)
|
|
|
frameshift
|
nucleotide groups change in composition
|
|
|
suppressed
|
when a first mutation's effect is blocked by the position of the second mutation
|
|
|
codon
|
nucleotide triplets
|
|
|
3 deletions or 3 insertions could do what
|
restore the function of a protein that have codons that are thre nucleotides long. NOT 4 NUCLEOTIDES B/C 3 DELETIONS WILL NOT RETURN TO BEGINNING.
|
|
|
STOP codon
|
UAA, UAG, UGA 3 of 64 codes are stop codons the others are for 20 amino acids
|
|
|
degenerate genetic code
|
different codons can code for the same amino acid there are 61 codons and they only make a total of 20 amino acids (about 3 different codons per amino acid)
|
|
|
tRNA charging
|
adding the amino acid to tRNA through the synthetase.
|
|
|
tRNA has a triplet of nucleotides called ________?
|
anticodon located in the anticodon loop
|
|
|
anticodons are complementary to?
|
codons of mRNA (except for stop codons)
|
|
|
aminoacyl-tRNA synthetase
|
separate synthetase for each amino acid - many synthetases needs to recognize multiple tRNA's
|
|
|
How many binding sites to synthetases have?
|
2 binding sites - one that contains an amino acid, and the other recognizes appropriate tRNA's
|
|
|
what is the complex that catalyzes protein synthesis?
|
ribosome
|
|
|
Ribosomes consist of how many units
|
2 larger subunit has 2 RNA molecules (3 if prokaryotic) and the smaller subunit has 1 RNA molecule
|
|
|
Ribosome facts
|
1. bring together the components needed for protein synthesis
2. add the amino acid attached to the charged tRNA to the growing protein chain A) aminoacyl - position that a charged tRNA enters the ribosome. a ribosome region called 'decoding center' ensures only the right charged tRNA is accepted. P) peptidyl - position that the amino acid of a charged tRNA is joined to the protein chain. A ribosome region called the peptidyl-transferase center' catalyzes this reaction E) exit- tRNA is ejected from the ribosome |
|
|
protein:enzyme
RNA: |
ribosome
|
|
|
prokaryotes mRNA translation
|
mRNA between the beginning of the transcription and the beginning of the gene called the 5' UTR contains a sequence that orientates the ribosome - called Shine Dalgarno
|
|
|
Initiation factors (IF)
|
promote the assembly of the complete ribosome
|
|
|
what is the usual first codon for the special initiator for formyl-methionine amino acid
|
AUG
|
|
|
mRNA must be processed and exported from the nucleus to the cytoplasm before translation can begin in what type of cell
|
eukaryotic cells
|
|
|
elongation factor Tu vs EF-G
|
a protein that brings charged tRNA to the A site of the ribosome while EF-G shifts the ribosome along the mRNA by one codon
|
|
|
RF
|
release factor
|
|
|
operators and activator sites
|
binding sites that influence the ability of RNA polymerase to bind and begin transcription for a given promoter
|
|
|
positive regulation
|
binding of some regulator proteins to activator binding sites and can increase RNA polymerase activity
|
|
|
negative regulation
|
binding of some regulator proteins to operators can decrease RNA polymerase activity
|
|
|
allosteric effector
|
signal molecule that binds to the allosteric site turning on or off gene expression
|
|
|
lac operon
|
model gene regulation
|
|
|
three genes that are active as enzymes are called
|
structural genes
|
|
|
LacI - negative regulation
|
inhibits gene expression by binding to operator and blockin the progress of RNA polymerase
|
|
|
CAP-cAMP
|
positive regulator
|
|
|
constitutive
|
always on, in reference to lac operon
|
|
|
cis acting
|
DNA operator only regulates genes on the same DNA molecule
|
|
|
trans acting
|
protein repressor regulates genes on any DNA molecule in the cell
|

