![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
78 Cards in this Set
- Front
- Back
|
Chap 12 # 1
What is semiconservative replication? |
In semiconservative replication, the original 2 strands of the double helix serve as templates for new strands of DNA. When replication is complete, two double-stranded DNA molecules will be present. Each will consist of one original template strand and one newly synthesized strand that is complementary to the template.
|
|
|
Chap 12 # 2
How did Meselson and Stahl demonstrate that replication in E. Coli takes place in a semiconservative manner? |
By growing E. Coli cells in a medium containing the heavy isotope of nitrogen (15 N) for several generations. The (15N) was incorporated into the DNA of the E. Coli cells. The E. Coli cells were then switched to a medium containing the common form of nitrogen (14N) and allowed to proceed through a few cycles of cellular generations. Samples of the bacteria were removed at each cellular generation. Using equilibrium density gradient centrifugation, they were able to distinguish DNA's that contained only (15N) from DNA's that contained only (14N) or a mixture of both because DNA's containing the (15N) isotope are heavier. The more (15N) a DNA molecule containes, the further it will sediment during equilibrium density gradient centrifugation. DNA from (15N)produced only a single band at expected position during centrifugation. After one round of replication in the (14N) medium and centrifugation 1 band was present at an intermediate to that of (15N) and after 2 rounds of replication 2 bands were present one at an intermediate to (15N) and one at the expected position for DNA only containing (14N). Results are consistent with predictions of semiconservative replication and incompatible with conservative and dispersive replication.
|
|
|
Chap 12 # 3
Draw a molecule of DNA undergoing Theta replication. On your drawing, identify 1) origin, 2) polarity 5' & 3' ends), 3) leading and lagging strands, 4) Okazaki Fragments, and 5) location of primers. |
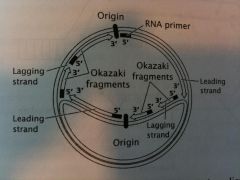
Draw this.
|
|
|
Chap 12 # 4
Draw a molecule of DNA undergoing rolling-circle replication. On your drawing, identify 1) origin, 2) polarity (5' & 3' ends) of all templae strands and newly synthesized strands, 3) leading and lagging strands, 4) Okazaki fragments, and 5) location of primers. |
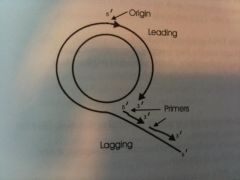
Draw this.
|
|
|
Chap 12 # 8
List the different proteins and enzymes taking part in bacterial replication. Give the function of each in the replication process. |
1) DNA polymerase III is the primary replication polymerase. It elongates a new nucleotide strand from the 3'-OH of the primer.
2) DNA polymerase I removes the RNA nucleotides of the primers and replaces them with DNA molecules. 3) DNA ligase connects Okazaki fragments by sealing nicks in the sugar phosphate backbone. 4) DNA primase synthesizes the RNA primers that provide the 3'-OH group needed for DNA polymerase III to initiate DNA synthesis. 5) DNA helicase unwinds the double helix by breaking the hydrogen bonding between the 2 strands at the replication fork. 6) DNA gyrase reduces DNA supercoiling and torsional strain that is created ahead of the replication fork by making double-stranded breaks in the DNA and passing another segment of the helix through the break before resealing it. Gyrase is also called topoisomerase II. 7) Initiator proteins bind to the replication origin and unwind short regions of DNA 8) Single-stranded binding protein (SSB protein) stabilizes single-stranded DNA prior to replication by binding to it, thus preventing the DNA from pairing with complementary sequences. |
|
|
CHAP 12 # 9
What similarities and differences exist in the enzymatic activities of DNA polymerase I, II, and III? What is the function of each type of DNA polymerase in bacterial cells? |
Each of the 3 DNA polymerases has a 5' to 3' polymerase activity. They differ in their exonuclease activities. DNA polymerase I has a 3' to 5' as well as a 5' to 3' exonuclease activity. DNA polymerase II & III have only a 3' to 5' exonuclease activity.
1) DNA Polymerase I carries out proofreading. It aslo removes and replaces the RNA primers used to initiate DNA synthesis. 2) DNA polymerase II functions as a DNA repair polymerase. It restarts replication after DNA damage has halted replication. It has proofreading activity. 3) DNA polymerase III is the primary replication enzyme and also has a proofreading function in replication. |
|
|
CHAP 12 # 10
Why is primase required for replication? |
Primase is a DNA-dependent RNA polymerase. Primase synthesizes the short RNA molecules, or primers, that have a free 3'-OH to which DNA polymerase can attach deoxyribonucleotides in replication initiation. The DNA polymerases require a free 3'-OH to which they add nucleotides, and therefore they cannot initiate replication. Primase does not have this requirement.
|
|
|
CHAP 12 # 11
What 3 mechanisms ensure the accuracy of replication in bacteria? |
1) Highly accurate nucleotide selection by the DNA polymerase when pairing bases.
2) the proofreading function of DNA polymerase, which removes incorrectly inserted bases. 3) a mismatch repair apparatus that repairs mistakes after replication is complete. |
|
|
CHAP 12 # 13
In what ways is eukaryotic replication similar to bacterial replication, and in what ways is it different? |
1) Semiconservative replication
2) replication origins serve as starting points for replication. 3) Short segments of RNA called primers provide a 3'-OH for DNA polymerases to begin synthesis of new strands. 4) Synthesis occurs in 5' to 3' direction. 5) The template strand is read in 3' to 5' direction. 6) Deoxyribonucleoside triphosphates are the substrates 7) Replication is continuous on the leading strand and discontinuous on the lagging strand. Eukaryotic DNA replication differs from bacterial replication in that: 1) It has multiple origins of replications per chromosome. 2) It has several different DNA polymerases with different functions. 3) Immediately following DNA replication, assembly of nucleosomes takes place. 1) It has mu |
|
|
CHAP 12 # 14
What is the end-of-chromosome problem for replication? Why, in the absence of telomerase, do the ends of chromosomes get progressively shorter each time the DNA is replicated? |
For DNA polymerases to work, they need the presence of a 3'-OH group to which to add a nucleotide. At the ends of the chromosomes when the RNA primer is removed, ther is no adjacent 3'-OH group to which to add a nucleotide, thus no nucleotides are added leaving a gap at the end of the chromosome. Telomerase can extend the single stranded protruding end by pairing with the overhanging 3' end of the DNA and adding a repeated sequence of nucleotides. In the absence of telomerase, DNA polymerase will be unable to add nucleotides to the end of the strand. After multiple rounds of replication without a functional telomerase the chromosome will become progressively shorter.
|
|
|
CHAP 12 # 21
A circular molecule of DNA contains 1 million base pairs. If DNA synthesis at a replication fork occurs at a rate of 100,000 nucleotides per minute, how long will theta replication require to completely replicate a molecule, assuming that theta replication is bi-directional? How long will replication of this circular chromosome take by rolling-circle replication? Ignore replication of the displaced strand in rolling-circle replication. |
In bi-directional replication there are 2 replication forks, each proceeding at a rate of 100,000 nucleotides per minute. Therefore it would require 5 minutes for circular DNA to be replicated by bi-directional replication because each fork could synthesize 500,000 nucleotides (5 min x 100,000 nucleotides per min) within the time period. Because rolling-circle replication is unidirectional and thus has only one replication fork, 10 minutes will be required to replicate the entire circular molecule.
|
|
|
CHAP 12 # 23
The following diagram represents a DNA molecule that is undergoing replication. Draw the strands of the newly synthesized DNA and identify the following: 1) Polarity of newly synthesized strands 2) Leading and Lagging strands 3) RNA primers. |
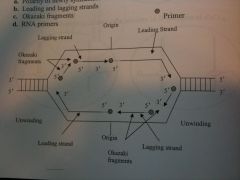
Draw This.
|
|
|
CHAP 12 # 24
What would be the effect on DNA replication of mutations that destroyed each of the following activities in DNA polymerase I? a) 3' - 5' exonuclease activity b) 5' - 3' exonuclease activity c) 5' - 3' polymerase activity |
A) The 3' - 5' exonuclease activity is important for proofreading newly synthesized DNA. If the activity is nonfunctional, then the fidelity of replication by DNA polymerase I will decrease, resulting in more misincorporated bases in the DNA
B) Loss of the 5' - 3' exonuclease activity would result in the RNA primers used to initiate replication not being removed by DNA polymerase I. C) DNA polymerase I would be unable to synthesize new DNA strands if the 5' - 3' polymerase activity was destroyed. RNA primers could be removed by DNA polymerase I using the 5' - 3' exonuclease activity, but new DNA sequencews could not be added in their place by DNA polymerase I. |
|
|
CHAP 12 # 25
How would DNA replication be affected in a cell that is lacking topoisomerase? |
Topoisomerase II (gyrase) reduces the positive supercoiling or torsional strain that develops ahead of the replication fork due to the unwinding of the double helix. if the topoisomerase activity was lacking, then the torsional strain would continue to increase, making it more difficult to unwind the double helix. Ultimately, the increasing strain would lead to an inhibition of the replication fork movement.
|
|
|
Chap 13 # 2
What are the major classes of cellular RNA? Where would you expect to find each class of RNA within eukaryotic cells? |
1) Ribosomal RNA, or rRNA, is found in the cytoplasm.
2) Transfer RNA, or tRNA, is found in the cytoplasm. 3) Messenger RNA, or mRNA, is found in the cytoplasm, (However, pre-mRNA is found only in the nucleus. 4) Small nuclear RNA, or smRNA, is found in the nucleus as part of riboproteins called snrps. 5) Small nucleolar RNA, or snoRNA, is found in the nucleus. 6) Small cytoplasmic RNA, or scRNA, is found in the cytoplasm. |
|
|
Chap 13 # 4
What parts of DNA make up a transcription unit? Draw and label a typical transcription unit in a bacterial cell. |

Draw This.
|
|
|
Chap 13 # 6
Describe the structure of RNA polymerase. |
Bacterial RNA polymerase consists of several polypeptides. The RNA polymerase core enzyme is composed of 4 polypeptide subunits: 2 copies of the alpha subunit, the beta subunit, and the beta prime subunit. The addition of a sigma factor to the core enzyme forms the RNA polymerase holoenzyme.
|
|
|
Chap 13 # 8
What are the three basic stages of transcription? Describe what happens at each. |
1) Initiation: Transcription proteins assemble at the promoter to form the basal transcription apparatus and begins synthesis of RNA.
2) Elongation: RNA polymerase moves along the DNA template in a 3' to 5' direction unwinding the DNA and synthesizing RNA in a 5' to 3' direction. 3) Termination: Synthesis of RNA is terminated, and the RNA molecule separates from the DNA template. |
|
|
Chap 13 # 9
Draw and label a typical bacterial promoter. Include any common consensus sequences. |

Draw This.
|
|
|
CHAP 13 # 11
How is the process of transcription in eukaryotic cells different from that in bacterial cells? |
Eukaryotic transcription requires the action of 3 RNA polymerases. Each type of polymerase recognizes and transcribes from different types of promoters. Binding to the promoter and initiation from the promoter requires the action of many protein factors; different promoters require different sets of protein factors. The RNA molecule produced by transcription in eukaryotic cells usually requires extensive processing, such as the addition of a 5' cap, a 3' poly(A) tail, and the removal of introns prior to becoming functional. Bacterial promoters tend to be more uniform in composition, and only 1 RNA polymerase does transcription. Bacterial RNAs are typically functional once transcription has taken place.
|
|
|
CHAP 13 # 16
The following diagram represents DNA that is part of the RNA-coding sequence of a transcription unit. The bottom strand is the template strand. Give the sequence found on the RNA molecule transcribed from this DNA and label the 5' and 3' ends of the RNA 5'-ATAGGCGATGCCA-3' 3'-TATCCGCTACGGT-5' |
The RNA molecule would be complementary to the template strand, contain uracil, and be synthesized in an antiparallel fasion.
The RNA strand contains the ssame sequence as the nontemplate DNA strand except that the RNA strand contains uracil instead of thymine. |
|
|
CHAP 13 # 19
Write the consensus sequence for the following set of nucleotide sequences. AGGAGTT AGCTATT TGCAATA ACGAAAA TCCTAAT TGCAATT |
The consensus sequence is identified by determining which nucleotide is used most frequently at each position. For the 2 nucleotides that occur at an equal frequency at the first position, both are listed at that position and identified by a slash mark:
T/A GCAATT |
|
|
CHAP 13 # 20
List at least 5 properties that DNA polymerases and RNA polymerases have in common. List at least 3 differences. |
Similarities:
1) Both use DNA templates 2) DNA templates are read in the 3' - 5' direction. 3) the complementary strand is synthesized in a 5' - 3' direction that is antiparallel to the template. 4) Both use triphosphates as substrates 5) Their actions are enhanced by accessory proteins Differences: 1) RNA polymerases use ribonucleoside triphosphates as substrates, whereas DNA polymerases use deoxyribonucleoside triphosphates 2) DNA polymerases require a primer that provides an available 3'-OH group where synthesis begins, whereas RNA polymerases do not require primers to begin synthesis 3) RNA polymerases synthesize a copy off only one of the DNA strands, Whereas DNA polymerases can synthesize copies of both strands. |
|
|
CHAP 14 # 2
What are some characteristics of introns? |
Eukaryotic genes commonly contain introns. However, introns are rare in bacterial genes. The number of introns found in an organism's genome is typically related to complexity - More complex organism's possess more introns. Introns typically do not encode proteins and are usually larger in size than exons.
|
|
|
CHAP 14 # 4
What are the 3 principal elements in mRNA sequences in bacterial cells? |
1) The 5' untranslated region, which contains the Shine-Dalgarno sequence
2) The protein encoding region 3) The 3' untranslated region |
|
|
CHAP 14 # 5
What is the function of the Shine-Delgarno consensus sequence? |
The Shine-Delgarno consensus sequence functions as the ribosome-binding site on the mRNA molecule.
|
|
|
CHAP 14 # 6
What is the 5' cap? |
The 5' end of eukaryotic mRNA is modified by the addition of the 5' cap. The cap consists of an extra guanine nucleotide linked to 5' to 5' to the mRNA molecule. This nucleotide is methylated at position 7 of the base. The ribose sugars of adjacent bases may be methylated at the 2'-OH.
|
|
|
Chap 14 # 7
How is the poly(A) tail added to Pre-mRNA? What is the purpose of the poly(A) Tail? |
Initially, a complex consisting of several proteins forms on the 3' UTR of the pre-mRNA molecule. Cleavage and polyadenylation specificity factor (CPSF) bind to the AAUAAA consensus sequence, which is located upstream of the 3' cleavage site. Another protein, cleavage stimulation factor (CsTF), binds downstream of the cleavage site. 2 cleavage factors (CFI & CFII) and polyadenylate polymerase (PAP) also become part of the complex. Once the complex has formed, the pre-mRNA is cleaved. CsTF and the 2 cleavage factors leave the complex. PAP adds approximately 10 adenine nucleotides to the 3' end of the pre-mRNA molecule. The addition of the short poly(A) tail allows for the binding of the poly(A) protein (PABII) to the tail. PABII increases the rate of polyadenylation, which subsequently allows for more PABII protein to bind to the tail.
|
|
|
Chap 14 # 8
What makes up the spliceosome? What is the function of the spliceosome? |
The spliceosome consists of 5 small ribonucleotides (snRNPs). Each snRNP is composed of multiple proteins and a single small nuclear RNA molecule or snRNA. The snRNPs are identified by which snRNA (U1, U2, U3, U4, U5, or U6) each contains. Splicing of pre-mRNA nuclear introns takes place within the spliceosome.
|
|
|
Chap 14 # 9
Explain the process of pre-mRNA splicing in nuclear genes. |
Removal of the intron from the pre-mRNA requires the assembly of the spliceosome complex on the pre-mRNA, cleavage at both the 5' & 3' splice sites of the intron, and 2 transesterification reactions ultimately leading to the joining of the 2 exons. Initially, snRNP UI binds to the 5' splice site through complementary base pairing of the UI snRNA. Next, snRNP U2 binds to the branch point within the intron. The U5 and U4-U6 complex joins the spliceosome, resulting in the looping of the intron so that the bracnch point and 5' splice site of the intron are now adjacent to each other. UI and U4 now disassociate from the spliceosome, and the spliceosome is activated. The pre-mRNA is then cleaved at the 5' splice site, producing an exon with a 3'-OH. The 5' end of the intron folds back and forms 5'-2' phosphodiester linkage through the first transesterification reaction with the adenine nucleotide at the branch point of the intron. This looped structure is called the lariat. Next, the 3' splice site is cleaved and then immediately ligated to the 3;-OH of the first exon through the second transesterification reaction. Thus, the exons are now joined and the intron has been excised.
|
|
|
CHAP 14 # 13
What are some of the modifications in tRNA that occur through processing? |
1) The precursor RNA may be cleaved into smaller molecules.
2) Nucleotides at both 5' & 3' ends of the tRNAs may be removed or trimmed. 3) Standard bases can be altered by base-modifying enzymes. |
|
|
CHAP 14 # 15
Explain how rRNA is processed. |
Most rRNAs are synthesized as large precursor RNAs that are processed by methylation, cleavage, and trimming to produce the mature mRNA molecules. In E. Coli, methylation occurs to specific bases and the 2'-OH of the ribose sugars of the 30S rRNA precursor. The 30S precursor is cleaved and trimmed to produce the 16S rRNA, 23S rRNA, and the 5S rRNA. In eukaryotes, a similar process occurs. However, small nucleolar RNAs help to cleave and modify the precursor rRNAs.
|
|
|
CHAP 14 # 16
What is the origin of siRNAs and microRNAs? What do these RNA molecules do in the cell? |
The siRNAs originate from the cleavage of mRNAs, RNA transposons, and RNA viruses by enzyme Dicer. Dicer may produce multiple siRNAs from a single double-stranded RNA molecule. The double-stranded RNA molecule may occur due to the formation of hairpins or by duplexes between different RNA molecules. The miRNAs arise from the cleavage of individual RNA molecules that are distinct from other genes. The enzyme Dicer cleaves these RNA molecules that have formed small hairpins. A single miRNA is produced from a single RNA molecule.
Both siRNAs and microRNAs silence gene expression through a process called RNA interference. Both function by shutting off gene expression of a cell's own genes or to shut off expression of genes from the invading foreign genes of viruses or transposons. The micrRNAs typically silence genes that are different from those which tha microRNAs are transcribed. However, the siRNAs usually silence genes from which they are transcribed. |
|
|
CHAP 14 # 20
How do the mRNA of bacterial cells and the pre-mRNA of eukaryotic cells differ? How do the mature mRNAs of bacterial cells and eukaryotic cells differ? |
Bacterial mRNA is translated immediately upon being transcribed. Eukaryotic pre-mRNA must be processed. Bacterial mRNA and eukaryotic pre-mRNA have similarities in structure. Each has a 5' untranslated region as well as a 3' untranslated region. Both also have protein-coding regions. However, the protein-coding region of the pre-mRNA is disrupted by introns. The eukaryotic pre-mRNA must be processed to produce the mature mRNA. Eukaryotic mRNA has a 5' cap and a poly(A) tail, unlike bacterial mRNAs. Bacterial mRNA also contains the Shine-Dalgarno consensus sequence. Eukaryotic mRNA does not have the equivalent.
|
|
|
CHAP 14 # 21
Draw a typical eukaryotic gene and the pre-mRNA and mRNA derived from it. Assume that the gene contains 3 exons. Identify the following items and, for each item, give a brief description of its function. a) 5' untranslated region b) Promoter c) AAUAAA consensus sequence d) Transcription start site. e) 3' untranslated region f) Introns g) Exons h) Poly(A) tail i) 5' Cap |
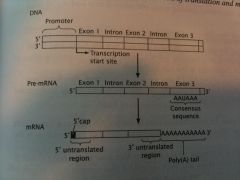
Draw This.
|
|
|
MY CARDS
What is the function of DNA Polymerase III? |
It is the primary replication polymerase. It elongates a new nucleotide strand from the 3'-OH of the primer.
|
|
|
MY CARDS
What is the function of DNA Polymerase I? |
It removes the RNA nucleotides of the primers and replaces them with DNA molecules.
|
|
|
MY CARDS
What is the function of DNA Ligase? |
It connects the Okazaki fragments by sealing nicks in the sugar phosphate backbone.
|
|
|
MY CARDS
What is the function of DNA Primase? |
It synthesizes the RNA primers that provide the 3'OH group needed for DNA polymerase III to initiate synthesis.
|
|
|
MY CARDS
What is the function of DNA Helicase? |
It unwinds the double helix by breaking the hydrogen bonding between the 2 strands at the replication fork.
|
|
|
MY CARDS
What is the function of DNA Gyrase? |
It reduces the supercoiling and torsional strain that is created ahead of the replication fork by making double-stranded breaks in the DNA and passing another segment of the helix through the break before resealing it. Gyrase also called topoismerase.
|
|
|
MY CARDS
What is the function of Initiator Proteins? |
They bind to the replication origin and unwind short regions of DNA.
|
|
|
MY CARDS
What is the function of Single-stranded binding protein (SSB)? |
It stabilizes single-stranded DNA prior to replication by binding to it, thus preventing the DNA from pairing with complementary sequences.
|
|
|
MY CARDS
Describe function of DNA polymerase I. |
Carries out proofreading. It also removes and replaces the RNA primers used to initiate DNA synthesis.
|
|
|
MY CARDS
Describe function of DNA polymerase II. |
Functions as a DNA repair polymerase. It restarts replication after DNA damage has halted replication. It has proofreading activity.
|
|
|
MY CARDS
Describe function of DNA polymerase III. |
It is the primary replication enzyme and also has a proofreading function in replication.
|
|
|
MY CARDS
What is the similarities of the 3 DNA polymerases? |
All have 5' to 3' polymerase activity.
|
|
|
MY CARDS
What are the differences between the 3 DNA polymerases? |
they differ in exonuclease activities.
DNA polymerase I & III have only a 3' to 5' exonuclease activity. DNA polymerase II has both 3' to 5' and 5' to 3' exonuclease activity. |
|
|
MY CARDS
Name the major classes of RNA. |
Ribosomal (rRNA)
Transfer (tRNA) Messenger (mRNA) Small Nuclear (smRNA) Small Nucleolar (snoRNA) Smal Cytoplasmic (scRNA) |
|
|
MY CARDS
Where is Ribosomal RNA found? |
In the cytoplasm.
|
|
|
MY CARDS
Where is Transfer RNA found? |
In the Cytoplasm.
|
|
|
MY CARDS
Where is Messenger RNA found? |
In the Cytoplasm. (However, pre-mRNA is only found in the nucleus)
|
|
|
MY CARDS
Where is pre-mRNA found? |
In the Nucleus.
|
|
|
MY CARDS
Where is Small nuclear RNA found? |
In the nucleus (as part of riboproteins called snrps.)
|
|
|
MY CARDS
Where is Small Nucleolar RNA found? |
In the Nucleus.
|
|
|
MY CARDS
Where is Small CytoplasmicRNA found? |
In the Nucleus.
|
|
|
MY CARDS
What are snrps? |
small nuclear ribosomal proteins.
|
|
|
MY CARDS
Describe Initiation. |
Transcription proteins assemble at the promoter to form the basal transcription apparatus and begins synthesis of RNA.
|
|
|
MY CARDS
Describe Elongation. |
RNA polymerase moves along the DNA template in a 3' to 5' direction unwinding the DNA and synthesizing RNA in a 5' to 3' direction.
|
|
|
MY CARDS
Describe Termination. |
Synthesis of RNA is terminated, and the RNA molecule seperates from the DNA template.
|
|
|
CHAP 15 # 3
What are isoaccepting tRNAs? |
Isoaccepting tRNAs are tRNA molecules that have different anticodon sequences but accept the same amino acids.
|
|
|
CHAP 15 # 4
What is the significance of the fact that many synonymous codons differ only in the third nucleotide position? |
Synonymous codons code for the same amino acid, or, in other words, have the same meaning. A nucleotide at the 3rd position of a codon pairs with a nucleotide in the 1st position of the anticodon. Unlike the other nucleotide positions involved in the codon-anticodon pairing, this pairing is often weak or wobbles, and nonstandard pairings can occur. Because of the wobble, or nonstandard base-pairing with the anticodons, affects the 3rd nucleotide position, the redundency of codons ensures that the correct amino acid is inserted in the protein when nonstandard pairings occur.
|
|
|
CHAP 15 # 5A
Define: Reading Frame. |
The reading frame refers to how the nucleotides in a nucleic acid molecule are grouped into codons containing 3 nucleotides. Each sequence of nucleotides has 3 possible sets of codons, or reading frames.
|
|
|
CHAP 15 # 5B
Define: Overlapping code. |
If an overlapping code is present, then a single nucleotide is included in more than 1 codon. The result for a sequence of nucleotides is that more than one type of polypeptide can be encoded within that sequence.
|
|
|
CHAP 15 # 5C
Define: Nonoverlapping code. |
In a nonoverlapping code, a single nucleotide is part of only 1 codon. This results inthe production of a single type of polypeptide from one polynucleotide sequence.
|
|
|
CHAP 15 # 5D
Define: Initiation codon: |
An initiation codon establishes the appropriate reading frame and specifies the first amino acid of the protein chain. Typically, the initiation codon is AUG; however, GUG and UUG can also serve as initiation codons.
|
|
|
CHAP 15 # 5E
Define: Termination codon: |
The termination codon signals the termination or end of translation and the end of the protein molecule. There are 3 types of termination codons - UAA, UAG, and UGA - which can also be referred to as stop codons or nonsense codons. These codons do not code for amino acids.
|
|
|
CHAP 15 # 5F
Define: Sense codon: |
A sense codon is a group of 3 nucleotides that code for an amino acid. There are 61 sense codons that code for 20 amino acids commonly found in proteins.
|
|
|
CHAP 15 # 5G
Define: Nonsense codon: |
Nonsense codons or termination codons signal the end of translation. These codons do not code for amino acids.
|
|
|
CHAP 15 # 5H
Define: Universal code: |
In a universal code, each codon specifies, or codes, for the same amino acid in all organisms. The genetic code is nearly universal, but not completely. Most of the exceptions occur in mitochondrial genes.
|
|
|
CHAP 15 # 5I
Define: Nonuniversal codons: |
Most codons are universal (or nearly universal) in that they specify the same amino acids in almost all organisms. However, there are exceptions where a codon has different meanings in different organisms. Most of the know exceptions are the termination codons, which in some organisms do code for amino acids. Occasionally, a sense codon is substituted for another sense codon.
|
|
|
CHAP 15 # 7
How are tRNAs linked to their corresponding amino acids? |
Each of the 20 different amino acids that are commonly found in proteins has a corresponding aminoacyl-tRNA synthetase that covalently links tha amino acid to the correct tRNA molecule.
|
|
|
CHAP 15 # 9
How does the process of initiation differ in bacterial cells and eukaryotic cells? |
Bacterial initiation of translation requires that sequences in the
16S rRna of the small ribosomal subunit bind to the mRNA at the ribosome binding site or Shine-Dalgarno sequence. The Shine-Dalgarno sequence is essential in placing the ribosome over the start codon (typically AUG). In eukaryotes, there is no Shine-Dalgarno sequence. The small ribosomal subunit recognizes the 5' cap of the eukaryotic mRNA with the assistance of initiation factors. Next, the ribosomal small subunit migrates along the mRNA scanning for the AG start codon. In eukaryotes, the start codon is located with a consensus sequence called the Kozak sequence (5' - ACCAUGG - 3'). transcription in eukaryotes also requires more initiation factors. |
|
|
CHAP 15 # 14
What are some types of posttranslational modification of proteins? |
Several different modifications can occur to a protein following translation. Frequently, the amino terminal methionine may be removed. Sometimes, in bacteria only the formyl group is cleaved from the N-formyl methionine, leaving a methionine at the amino terminal. More extensive modification occurs in some proteins that are originally synthesized as precursor proteins. These precursor proteins are cleaved and trimmed by protease enzymes to produce a functionalprotein. Glycoproteins are produced by the attachment of carbohydrates to newly synthesized proteins. Molecular chaperones are needed by many proteins to ensure that the proteins are folded correctly. Secreted proteins that are targeted for the membrane or other cellular locations frequently have 15 to 30 amino acids, called the signal sequence, removed from the amino terminal. Finally acetylation of amino acids in the amino terminal of some eukaryotic proteins also occurs.
|
|
|
CHAP 15 # 15
Explain how some antibiotics work by affecting the process of protein synthesis. |
A number of antibiotics bind the ribosome and inhibit protein synthesis at different steps in translation. Some antibiotics, such as streptomycin, bind to the small subunit and inhibit translation initiation. Other antibiotics, such as chloramphenicol bind to the large subunit and block elongation of the peptide by preventing peptide bond formation. Antibiotics such as tetracycline and neomycin, bind the ribosome near the "A" site yet have different effects. Tetracyclines block entry of charged tRNAs to the "A" site, while neomycin induces translation errors. Finally, some antibiotics such as erythromycin, block the translation of the ribosome along the mRNA.
|
|
|
What 3 different methods were used to help break the genetic code? What did each reveal and what were the advantages and disadvantages?
|
Marshal Nirenberg and Johann Heinrich Matthaei used the enzyme polynucleotide kinase to create homopolymers of synthetic RNAs. Using a cell-free protein synthesizing system they were able to determine the amino acid coded by each homopolymer. By this method, the meanings for the amino acids specified by the codons UUU, AAA, CCC, and GGG were determined. The disadvantage is that the meanings for only 4 codons could be determined.
The same system was also used to create copolymers that contained random mixtures of 2 nucleotides in a known ratio. Different amino acids in the protein depended on the ratio of the 2 nucleotides. To determine or predict the composition of the codons, the frequency of amino acids produced using the copolymer was compared with the theoretical frequencies expected for the codons. A disadvantage of this procedure is that it depended on random incorporation of nucleotides, which did not always happen. A further problem was that the base sequence of the codon could not be determined - only the bases contained within the codon. The redundancy of the code also provided difficulties because several different codons could specify the same amino acid. To solve these problems, they mixed ribosomes bound to short RNAs of known sequences with charged tRNAs. The mixture was passed through a nitrocellulose filter to which the tRNAs paired to ribosome-mRNA stuck. They next determined the amino acids attached to the bound tRNAs. More then 50 codons were identified by this method. The difficulty is that not all tRNAs and codons could be identified with this method. Gobind Khorana and his colleagues used a 3rd method. They synthesized RNA molecules of known repeating sequences. Using a cell-free protein synthesizing system they produced proteins of alternating amino acids. However, this procedure could not specify which codon encodes which amino acid. |
|
|
CHAP 15 # 27
Arrange the following components of translation in the appropriate order in which they would appear or be used during protein synthesis. 70S initiation complex release factor 1 peptidyl transferase elongation factor G 30S initiation complex elongation factor TU initiation factor 3 fMet-tRNA |
fMet-tRNA
30S initiation complex Initiation factor 3 70S initiation complex Elongation factor Tu Peptidyl transferase elongation factor G Release factor 1 |
|

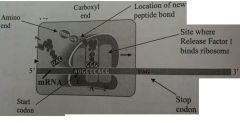
The following diagram illustrates a step in the process of translation.
Identify the following: a) 5' and 3' ends of the RNA b) A,P, and E sites c) Start codon d) Stop codon e) Amino and carboxyl ends of the newly synthesized polypeptide chain. f) Approximate location of the next peptide bond that will be formed g) Place on the ribosome where release factor 1 will bind |
|

