![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
123 Cards in this Set
- Front
- Back
|
How is DNA organized in the nucleus?
|
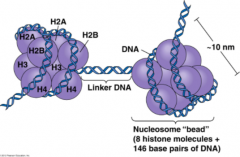
Condensed, chromatin form
- Negatively charged DNA loops twice around positively charged histone octamer to form nucleosome "bead" - Linker DNA forms the string |
|
|
What are the characteristics of Histones? What do they consist of? Organization?
|
- Rich in amino acids lysine and arginine
- H1 ties nucleosome beads together in a string (only histone not in the nucleosome core) - Nucleosome core consists of H2A, H2B, H3, H4 (each x2) |
|
|
What is the difference between heterochromatin and euchromatin?
|
- HeteroChromatin = Highly Condensed, transcriptionally inactive, sterically inaccessible
- Euchromatin = less condensed, transcriptionally active, sterically accessible ("Eu" = True, truly transcribed) |
|
|
Which DNA bases can be methylated? Function?
|
- Template strand Cytosine and Adenine are methylated during DNA replication
- Allows mismatch repair enzymes to distinguish between old and new strands in prokaryotes |
|
|
How can histones be activated and inactivated?
|
- Histone methylation inactivates transcription of DNA (Methylation makes DNA Mute)
- Histone acetylation relaxes DNA coiling, allowing for transcription of DNA (Acetylation makes DNA Active) |
|
|
What are the types of nucleotides?
|
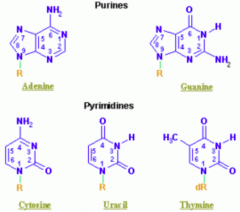
- Purines: A, G (2 rings) = PURe As Gold
- Pyrimidines: C, T, U (1 ring) = CUT the PY (pie) |
|
|
Which purine base has a ketone?
|
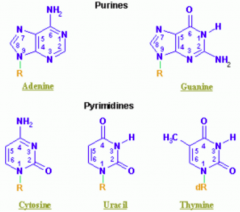
Guanine
|
|
|
Which nucleotide base has a methyl?
|
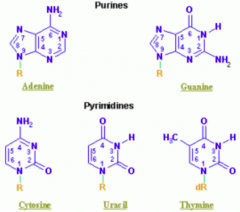
Thymine (THYmine has a meTHYl)
|
|
|
Which nucleotide base is formed by deamination of cytosine?
|
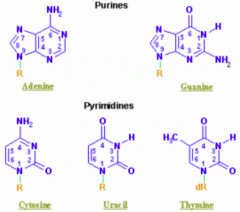
Uracil
|
|
|
Which nucleotide base is only found in RNA? Only found in DNA?
|
- RNA: uracil
- DNA: thymine |
|
|
What are the characteristics of nucleotide binding? Implications of more of one type of bond?
|
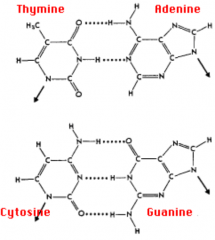
- G-C bond: 3 H bonds (stronger)
- A-T bond: 2 H bonds (weaker) - ↑ G-C content → ↑ melting temperature |
|
|
Which amino acids are necessary for purine synthesis?
|
GAG:
- Glycine - Aspartate - Glutamine |
|
|
What is the difference between a nucleoside and nucleotide?
|
- NucleoSide: base + ribose (Suger)
- NucleoTide: base + ribose + phosphaTe (linked by 3'-5' phosphodiester bond) |
|
|
How are purines synthesized de novo?
|
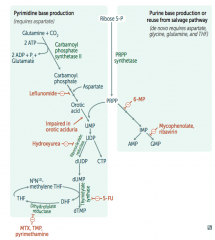
Right side:
- Start with sugar (Ribose-5P) + phosphate (PRPP) - Add base |
|
|
How are pyrimidines synthesized de novo?
|
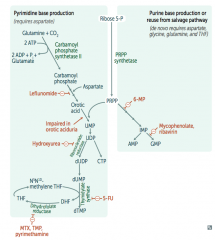
Left side:
- Make temporary base (eg, ribonucleotides) - Add sugar + phosphate (PRPP) - Modify base (eg, deoxyribonucleotides) |
|
|
How do ribonucleotides get converted to deoxyribonucleotides?
|
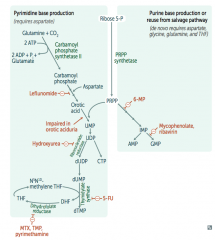
Ribonucleotide Reductase
|
|
|
Which drugs interfere with purine synthesis?
|
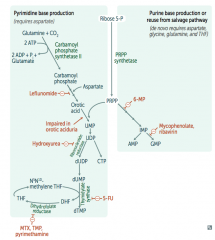
- Leflunomide
- Mycophenolate and Rivarin - Hydroxyurea - 6-Mercaptopurine (6-MP) and Azathioprine - 5-Fluorouracil (5-FU) - Methotrexate (MTX), Trimethoprim (TMP), and Pyrimethamine |
|
|
What is the mechanism of Hydroxyurea?
|
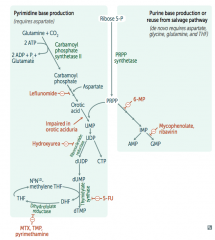
Interferes with purine synthesis:
- Inhibits Ribonucleotide Reductase |
|
|
What is the mechanism of 6-Mercaptopurine (6-MP)? Azathioprine?
|
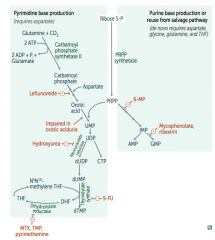
Interferes with purine synthesis:
- Blocks de novo purine synthesis Azathioprine is the prodrug |
|
|
What is the mechanism of 5-Fluorouracil (5-FU)?
|
![Interferes with purine synthesis:
- Inhibits thymidylate synthase (↓ deoxythmidine monophosphate [dTMP])](https://images.cram.com/images/upload-flashcards/40/26/71/4402671_m.png)
Interferes with purine synthesis:
- Inhibits thymidylate synthase (↓ deoxythmidine monophosphate [dTMP]) |
|
|
What is the mechanism of Methotrexate (MTX)?
|
![Interferes with purine synthesis:
- Inhibits dihydrofolate reductase (↓ deoxythmidine monophosphate [dTMP])](https://images.cram.com/images/upload-flashcards/40/26/65/4402665_m.png)
Interferes with purine synthesis:
- Inhibits dihydrofolate reductase (↓ deoxythmidine monophosphate [dTMP]) |
|
|
What is the mechanism of Trimethoprim (TMP) and Pyrimethamine?
|
![Interferes with purine synthesis:
- Inhibits dihydrofolate reductase (↓ deoxythmidine monophosphate [dTMP])
- Trimethoprim (TMP): bacteria
- Pyrimethamine: protozoa](https://images.cram.com/images/upload-flashcards/40/26/62/4402662_m.png)
Interferes with purine synthesis:
- Inhibits dihydrofolate reductase (↓ deoxythmidine monophosphate [dTMP]) - Trimethoprim (TMP): bacteria - Pyrimethamine: protozoa |
|
|
Which drug inhibits thymidylate synthase? Implications?
|
![5-Fluorouracil (5-FU)
- ↓ deoxythmidine monophosphate [dTMP]
- Interferes with purine synthesis](https://images.cram.com/images/upload-flashcards/40/26/59/4402659_m.png)
5-Fluorouracil (5-FU)
- ↓ deoxythmidine monophosphate [dTMP] - Interferes with purine synthesis |
|
|
Which drug inhibits dihydrofolate reductase? Implications?
|
![Methotrexate (MTX) and Trimethoprim (TMP)
- TMP inhibits bacterial form
- ↓ deoxythmidine monophosphate [dTMP]
- Interferes with purine synthesis](https://images.cram.com/images/upload-flashcards/40/26/56/4402656_m.png)
Methotrexate (MTX) and Trimethoprim (TMP)
- TMP inhibits bacterial form - ↓ deoxythmidine monophosphate [dTMP] - Interferes with purine synthesis |
|
|
Which drug inhibits ribonucleotide reductase? Implications?
|
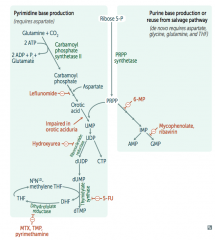
Hydroxyurea
- Interferes with purine synthesis |
|
|
Which drug blocks de novo purine synthesis? Implications?
|
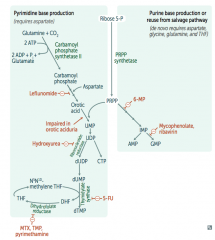
6-Mercaptopurine (6-MP)
- Interferes with purine synthesis |
|
|
What is the mechanism of:
- Leflunomide - Mycophenolate and Rivarin - Hydroxyurea - 6-Mercaptopurine (6-MP) and Azathioprine - 5-Fluorouracil (5-FU) - Methotrexate (MTX), Trimethoprim (TMP), and Pyrimethamine |
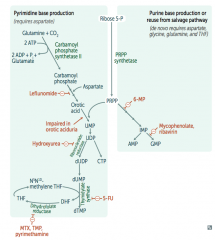
- Leflunomide: inhibits dihydroorotate dehydrogenase
- Mycophenolate and Ribavirin: inhibits IMP dehydrogenase - Hydroxyurea: inhibits ribonucleotide reductase - 6-Mercaptopurine (6-MP) and pro-drug Azathioprine: blocks de novo purine synthesis - 5-Fluorouracil (5-FU): inhibits thymidylate synthase (↓dTMP) - Methotrexate (MTX): inhibits dihydrofolate reductase (↓dTMP) - Trimethoprim (TMP): inhibits bacterial dihydrofolate reductase (↓dTMP) - Pyrimethamine: inhibits protozoan dihydrofolate reductase (↓dTMP) |
|
|
What is the function of Carbamoyl Phosphate?
|
Involved in 2 metabolic pathways:
- De novo pyrimidine synthesis - Urea cycle |
|
|
What can lead to an accumulation of Carbamoyl Phosphate? Implications?
|
- Ornithine Transcarbamoylase deficiency (OTC) - key enzyme in urea cycle
- Accumulation of Carbamoyl Phosphate is then converted to Orotic Acid |
|
|
What causes Orotic Aciduria?
|
- Inability to convert Orotic Acid to UMP (de novo pyrimidine synthesis pathway) because of a defect in UMP synthase (bifunctional enzyme)
- Autosomal recessive |
|
|
What are the findings of Orotic Aciduria?
|
- ↑ Orotic acid in the urine
- Megaloblastic anemia (no improvement w/ B12 or folic acid) - Failure to thrive * No hyperammonemia |
|
|
What are the findings of Ornithine Transcarbamoylase (OTC) deficiency, which leads to increased Orotic Acid?
|
- ↑ Orotic acid in the urine
- Megaloblastic anemia (no improvement w/ B12 or folic acid) - Failure to thrive * Hyperammonemia |
|
|
How do you treat Orotic Aciduria?
|
Oral uridine administration (because you are unable to convert orotic acid to UMP)
|
|
|
What are the purine salvage deficiencies?
|
- Severe Combined Immunodeficiency Disease (SCID): Adenosine Deaminase Deficiency
- Lesch-Nyhan Syndrome: defective purine salvage |
|
|
What is SCID? What causes it?
|
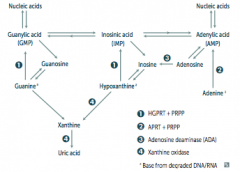
SCID = Severe Combined Immunodeficiency Disease
- Adenosine Deaminase (ADA) Deficiency - Autosomal recesive |
|
|
What are the implications of an Adenosine Deaminase (ADA) Deficiency?
|
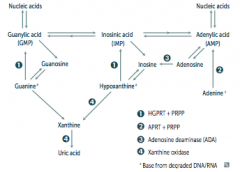
- Excess ATP and dATP imbalances nucleotide pool via feedback inhibition of ribonucleotide reductase
- This prevents DNA synthesis and ↓ lymphocyte count * One of the major causes of SCID (autosomal recessive) |
|
|
How do you treat Severe Combined Immunodeficiency Disease (SCID)?
|
Human gene therapy (replace Adenosine Deaminase (ADA))
|
|
|
What is Lesch-Nyhan Syndrome? What causes it?
|
- Syndrome that causes: HGPRT =
*Hyperuricemia *Gout *Peeved off (aggresion and self-mutilation) *Reta*rdation (intellectual disability) *dysTonia - Cause: defective purine salvage d/t absence of HGPRT, X-linked recessive |
|
|
What are the implications absent HGPRT? How do you treat?
|
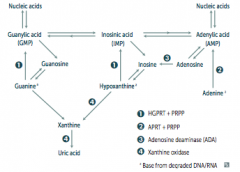
- HGPRT usually converts hypoxanthine to IMP and guanine to GMP
- Results in excess uric acid production and de novo purine synthesis - Lesch-Nyhan Syndrome He's Got Purine Recovery Trouble Treat: Allopurinol or Febuxostat (2nd line) |
|
|
What are the symptoms of Lesch-Nyhan Syndrome?
|
HGPRT =
*Hyperuricemia *Gout *Peeved off (aggresion and self-mutilation) *Reta*datoin (intellectual disability) *dysTonia (chorea) |
|
|
What are the features of the genetic code?
|
- Unambiguous
- Degenerate / Redundant - Commaless, Non-overlapping - Universal |
|
|
What does it mean that the genetic code is unambiguous?
|
Each codon specifies only one amino acid
|
|
|
What does it mean that the genetic code is degenerate / redundant?
|
- Most amino aids are coded by multiple codons
- Exceptions: methionine and tryptophan encoded by only 1 codon (AUG and UGG respectively) |
|
|
What does it mean that the genetic code is commaless, non-overlapping?
|
- Read from a fixed starting point as a continuous sequence of bases
- Exception: some viruses |
|
|
What does it mean that the genetic code is universal?
|
- Genetic code is conserved throughout evolution
- Exception: mitochondria in humans |
|
|
What are the types of point mutations in DNA from most severe to least severe?
|
Most severe
- Frameshift - Nonsense - Missense - Silent Least severe |
|
|
What happens in a frameshift mutation?
|
- Change resulting in misreading of all nucleotides downstream, usually resulting in a truncated, nonfunctional protein
- Most severe |
|
|
What happens in a nonsense mutation?
|
Change results in an early STOP codon
STOP the NONSENSE |
|
|
What happens in a missense mutation?
|
- Changed amino acid
- If conservative, new amino acid is similar in chemical structure |
|
|
What happens in a silent mutation?
|
- Same amino acid
- Often due to base change in 3rd position of codon (tRNA wobble) - Least severe |
|
|
What are the characteristics of DNA replication in prokaryotes and eukaryotes?
|
- Semi-conservative
- Continuous and discontinuous (Okazaki fragment) synthesis |
|
|
What is the term for the particular consensus sequence of base pairs in the genome where DNA replication begins? How does this compare for prokaryotes and eukaryotes?
|
Origin of Replication
- Prokaryotes: single - Eukaryotes: multiple |
|
|
What is the term for the Y-shaped region along the DNA template where leading and lagging strands are synthesized?
|
Replication Fork
|
|
|
What is the enzyme that unwinds the DNA template at the Replication Fork?
|
Helicase
|
|
|
What are the structures that prevent DNA strands from reannealing during DNA replication?
|
Single-Stranded Binding Proteins
|
|
|
What is the enzyme that creates a nick in the helix to relieve supercoils created during replication?
|
DNA Topoisomerases
|
|
|
Which drug inhibits the action of DNA Topoisomerases? Normal function?
|
- Fluoroquinolones inhibit DNA gyrase (prokaryotic topoisomerase II)
- Topoisomerases create nicks in the helix to relieve supercoils during DNA replication |
|
|
What is the enzyme that makes an RNA primer on which DNA polymerase III can initiate DNA replication?
|
Primase
|
|
|
What is the enzyme that is only found in prokaryotes and elongates the leading strand by adding deoxynucleotides to the 3' end, as well as elongates the lagging strand until it reaches the primer of preceding fragment?
|
DNA Polymerase III
|
|
|
What is the function of DNA Polymerase III?
|
- Prokaryotes only
- Elongates leading strand by adding deoxynucleotides to the 3' end (5'-3' direction) - Elongates lagging strand until it reaches the primer of preceding fragment (5'-3' direction) - 3'-5' exonuclease activity "proofreads" each added nucleotide (3'-5' direction) |
|
|
What proofreads the DNA replication of DNA Polymerase III?
|
3'-5' exonuclease
|
|
|
What is the enzyme that is only found in prokaryotes and degrades the RNA primer to replace it with DNA?
|
DNA Polymerase I
|
|
|
What is the function of DNA Polymerase I?
|
- Same functions as DNA Polymerase III (elongates leading and lagging strands in 5'-3' direction)
** Also degrades RNA primer with 5'-3' exonuclease activity to replace it with DNA |
|
|
What is the enzyme that catalyzes the formation of the phosphodiesterase bond within a strand of dsDNA (ie, joins Okazaki fragments)?
|
DNA Ligase (seals)
|
|
|
What is the enzyme that adds DNA to 3' ends of chromosomes to avoid loss of genetic material with every duplication?
|
Telomerase
|
|
|
What are the mechanisms of single strand DNA repair?
|
- Nucleotide excision repair
- Base excision repair - Mismatch repair |
|
|
What is the mechanism of nucleotide excision repair? Example?
|
- Specific endonucleases release the oligonucleotide-containing damaged bases
- DNA polymerase and ligase fill and reseal the gap - Repairs bulky helix distorting lesions Examples: - Defective in xeroderma pigmentosum, which prevents repair of pyrimidine dimers because of UV light exposure |
|
|
What is the mechanism of base excision repair?
|
- Base-specific glycosylase recognizes altered base and creates AP site (apurinic / apyrimidinic)
- One or more nucleotides are removed by AP-endonuclease, which cleaves the 5' end - Lyase cleaves the 3' end - DNA polymerase-β fills the gap and DNA ligase seals it - Important in repair of spontaneous / toxic deamination |
|
|
What is the mechanism of mismatch repair? Example?
|
- Newly synthesized strand is recognized
- Mismatched nucleotides are removed - Gap is filled and resealed Example: - Defective in hereditary nonpolyposis colorectal cancer (HNPCC) |
|
|
Which type of DNA repair fixes bulky helix distorting lesions?
|
Nucleotide Excision Repair
- Specific endonucleases release the oligonucleotide-containing damaged bases - DNA polymerase and ligase fill and reseal the gap |
|
|
Which type of DNA repair is important for repair of spontaneous / toxic deamination?
|
Base Excision Repair
- Specific glycosylases recognize and remove damaged bases - Apurine / apyrimidine endonuclease cuts DNA at both apurinic and apyrimidinic sites - Empty sugar is removed and the gap is filled and resealed |
|
|
What is an example of mutated nucleotide excision repair?
|
Xeroderma Pigmentosum
- Prevents repair of pyrimidine dimers because of UV light exposure |
|
|
What is an example of mutated mismatch repair?
|
Hereditary Nonpolyposis Colorectal Cancer (HNPCC)
|
|
|
What is the mechanism for double stranded DNA repair?
|
Non-homologous and joining
- Brings together 2 ends of DNA fragments to repair double-stranded breaks - No repairment for homology |
|
|
What is caused by double strand non-homologous and joining DNA repair?
|
Ataxia Telangiectasia
|
|
|
What is on the 5' end of the incoming nucleotide during DNA/RNA synthesis? Importance?
|
5' end contains a triphosphate (energy source for bond)
|
|
|
Drugs that block DNA replication often have what structure? Why?
|
- Modified 3' OH
- This prevents addition of the next nucleotide ("chain termination") |
|
|
What are the types of RNA?
- Most abundant? - Longest? - Smallest? |
- Most abundant: rRNA
- Longest: mRNA - Smallest: tRNA Rampant, Massive, Tiny |
|
|
What is the mRNA start codon? What does it encode?
|
AUG (or rarely GUG)
- Eukaryotes: codes for methionine, which may be removed before translation is complete - Prokaryotes: codes for formylmethionine (f-met) * AUG inAUGurates protein synthesis |
|
|
What are the mRNA stop codons?
|
- UGA (U Go Away)
- UAA (U Are Away) - UAG (U Are Gone |
|
|
What parts of the gene are important for regulation of gene expression?
|
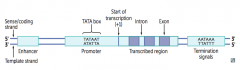
- Promoter
- Enhancer - Silencer |
|
|
What happens at the Promoter?
|
Site where RNA polymerase and multiple other transcription factors bind to DNA upstream from gene locus (AT-rich upstream sequence with TATA and CAAT boxes)
|
|
|
What happens if there are mutations in the gene promoter?
|
Dramatic decrease in amount of gene transcribed
|
|
|
What happens at an Enhancer?
|
Binds transcription factors to alter gene expression
|
|
|
What happens at a Silencer?
|
Binds negative regulators (repressors)
|
|
|
Where can enhancers and silencers be located?
|
Close to, far from, or even within (in an intron) of the gene whose expression they regulate
|
|
|
What are the types of RNA polymerases in eukaryotes? What do they make?
|
- RNA Polymerase I: makes rRNA (rampant)
- RNA Polymerase II: makes mRNA (massive) - RNA Polymerase III: makes tRNA (tiniest) |
|
|
What are the characteristics of all RNA Polymerases in eukaryotes?
|
- Can initiate chains
- No proofreading function - RNA Pol II can open DNA at promoter site |
|
|
Which toxin can inhibit RNA polymerase II? Implications?
|
- α-Amanitin (found in Amanita phalloides - a death cap mushroom)
- Causes severe hepatotoxicity if ingested |
|
|
What are the types of RNA polymerases in prokaryotes? What do they make?
|
Only 1 RNA polymerase (multi-subunit complex) - makes all three kinds of RNA (rRNA, mRNA, and tRNA)
|
|
|
What is the initial transcript of RNA called? What is it called when it is destined for translation?
|
- Heterogenous nuclear RNA (hrRNA) is the initial transcript
- pre-mRNA is destined for translation |
|
|
What are the methods of RNA processing in eukaryotes? Where does it take place? Result?
|
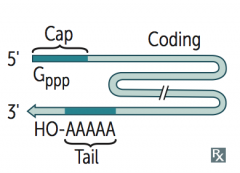
Takes place in nucleus:
- Capping on 5' end (addition of 7-methylguanosine cap) - Polyadenylation on 3' end (~200 A's) - Splicing out of introns Capped, tailed, and spliced transcript is called mRNA; only processed RNA is transported out of the nucleus |
|
|
What happens to the 5' end of pre-mRNA during processing in the nucleus?
|
Capping on 5' end (addition of 7-methylguanosine cap)
|
|
|
What happens to the 3' end of pre-mRNA during processing in the nucleus?
|
Polyadenylation (~200 A's); Poly-A polymerase does not require a template
AAUAA = polyadenylation signal |
|
|
How is pre-mRNA spliced?
|
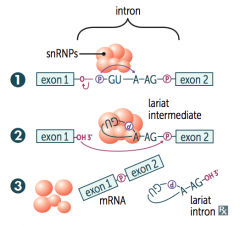
1. Primary transcript combines with snRNPs and other proteins to form spliceosome
2. Lariat-shaped (looped) intermediate is generated 3. Lariat is released to remove intron precisely and join 2 exons |
|
|
In what disease do patients make antibodies to spliceosomal snRNPs (involved in the splicing of pre-mRNAs)?
|
Lupus
|
|
|
What is the difference between introns and exons?
|
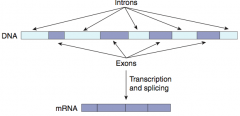
- Exons: contain genetic information coding for protein, they Exit the nucleus and are Expressed
- Introns: Intervening non-coding segments of DNA, that stay In the nucleus |
|
|
After splicing, what is the mRNA made of?
|
Exons (all of the introns have been removed)
|
|
|
What is the size and structure of tRNAs?
|
- 75-90 nucleotides
- 2° structure makes cloverleaf form - Anti-codon end is opposite 3' aminoacyl end |
|
|
Which end of the tRNA carries the amino acids? What code holds on to the amino acids?
|
- CCA at 3' end along with a high percentage of chemically modified bases
- AA is covalently bound to the 3' end of the tRNA CCA: Can Carry Amino acids |
|
|
What happens to the tRNA before and after binding an amino acid?
|
- Aminoacyl-tRNA synthetase scrutinizes AA before and after it binds to tRNA
- If incorrect, bond is hydrolyzed - The AA-tRNA bond has energy for formation of a peptide bond - A mischarged tRNA reads usual codon but inserts wrong amino acid |
|
|
What is necessary for accurate base pairing with tRNA?
|
- Only the first two nucleotide positions of an mRNA codon need to be accurate to bind
- Codons that differ in the 3rd "wobble" position may code for the same tRNA / amino acid - This is a result of degeneracy of genetic code |
|
|
What are the steps of protein synthesis?
|
1. Initiation
2. Elongation 3. Termination |
|
|
How does protein synthesis get initiated (step 1 of translation)?
|
- Initiation factors are activated by GTP hydrolysis
- IFs help assemble 40s ribosomal subunit with the initiator tRNA - IFs are released when the mRNA and ribosomal subunit assemble with the complex |
|
|
What are the ribosomal subunits in eukaryotes? Prokaryotes?
|
- Eukaryotoes (Evens): 40S + 60S → 80S
- PrOkaryotes (Odds): 30S + 50S → 70S |
|
|
How does protein synthesis get elongated (step 2 of translation)?
|
1. Aminoacyl-tRNA binds to A site (except for initiator methionine)
2. Ribosomal rRNA ("ribozyme") catalyzes peptide bond formation, transfers growing polypeptide to amino acid in A site 3. Ribosome advances 3 nucleotides toward 3' end of mRNA moving peptidyl tRNA to P site (translocation) |
|
|
What are the three sites on the ribosome for amino acids?
|
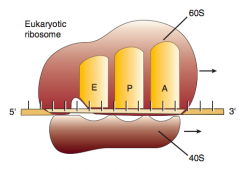
A: incoming Aminoacyl-tRNA
P: accommodates growing Peptide E: holds Empty tRNA as it Exits |
|
|
What happens in the first step of elongation (2nd step of protein synthesis)?
|
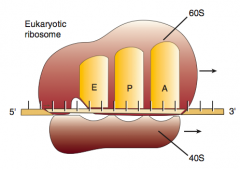
1. Aminoacyl-tRNA binds to A site (except for initiator methionine)
|
|
|
What happens in the second step of elongation (2nd step of protein synthesis), after Aminoacyl-tRNA binds to A site?
|
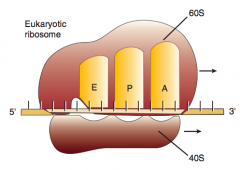
2. Ribosomal rRNA ("ribozyme") catalyzes peptide bond formation, transfers growing polypeptide to amino acid in A site
|
|
|
What happens in the third step of elongation (2nd step of protein synthesis), after Ribosomal rRNA ("ribozyme") catalyzes peptide bond formation?
|
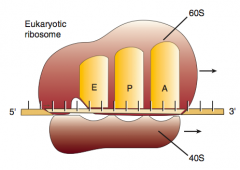
3. Ribosome advances 3 nucleotides toward 3' end of mRNA moving peptidyl tRNA to P site (translocation)
|
|
|
How does protein synthesis get terminated (step 3 of translation)?
|
Stop codon is recognized by Release Factor and completed protein is released from the ribosome
|
|
|
What are the types of antibiotics that affect protein synthesis?
|
- Aminoglycosides
- Tetracyclines - Chloramphenicol - Macrolides |
|
|
What is the mechanism of aminoglycosides?
|
Bind prokaryotic 30S subunit, which inhibits formation of initiation complexes and cause misreading of mRNA
|
|
|
What is the mechanism of tetracyclines?
|
Bind prokaryotic 30S subunit, which blocks aminoacyl tRNA from entering the acceptor site
|
|
|
What is the mechanism of chloramphenicol?
|
Binds prokaryotic 50S subunit and inhibits peptidyl transferase
|
|
|
What is the mechanism of macrolides?
|
Binds prokaryotic 50S subunit and prevents release of uncharged tRNA after it has donated its amino acid
|
|
|
Which antibiotics bind to the 30S subunit? Action?
|
- Aminoglycosides: inhibits formation of initiation coplexes and causes misreading of mRNA
- Tetracyclines: blocks aminoacyl tRNA from entering the acceptor site |
|
|
Which antibiotics bind to the 50S subunit? Action?
|
- Chloramphenicol: inhibits peptidyl transferase
- Macrolides: prevents release of uncharged tRNA after it has donated its AA |
|
|
What are the forms of post-translational modification?
|
- Trimming
- Covalent alterations - Proteasomal degradation |
|
|
What happens in "trimming" (a type of post-translational modification)?
|
Removal of N- or C-terminal propetides from zymogens to generate mature proteins
|
|
|
What happens in "covalent alterations" (a type of post-translational modification)?
|
Phosphorylation, glycosylation, hydroxylation, methylation, and acetylation
|
|
|
What happens in "proteasomal degradation" (a type of post-translational modification)?
|
Attachment of ubiquitin to defective proteins to tag them for breakdown
|
|
|
What is the function of chaperone proteins?
|
- Intracellular protein involved in facilitating and/or maintaining protein folding
- In yeast, some are heat shock proteins (eg, Hsp60) that are expressed at high temperature to prevent protein denaturing / misfolding |

