![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
37 Cards in this Set
- Front
- Back
|
DNA sites are mostly electrophillic or nusleophilllic?
|
nucleophillic
|
|
|
What is the source of most human mutations?
|
sperm cells, because they replicate ca. 40 times before they mature
|
|
|
Rare imino form of cytosine structure
- what nu does it bind to? |
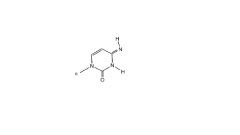
binds to Adenosine instead of Guadosine
|
|
|
Rare enol form of thymidine structure
what Nu does it bind to? |
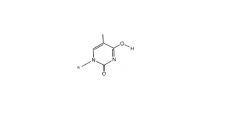
binds to G instead of A
|
|
|
Rare imino form of Adenosine structure
what Nu does it bind to? |
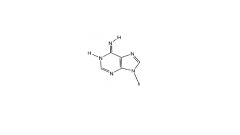
binds to C instead of T/U
|
|
|
Rare enol form of Guanosine structure
what Nu does it bind to? |
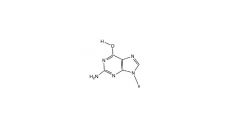
binds to T instead of C
|
|
|
Tautomeric shift mutation
- what happens - how is it repaired? |
a nucleotide may spontaneously transition to its rare tautomer form, then pair with the incorrect W-C base pair
- |
|
|
transition
transversion |
transition- purine to purine or pyrimidine to pyrimidine shift
transversion- purine to pyrimidine or pyrimidine to purine shift |
|
|
enolization
|
misspairing of the opposite base
- usual cause of transitions |
|
|
spontaneous deletion
|
at times, DNA is not a strict double helix
polymerase may skip a looped out sequence to make a spontaneous deletion - may occur when identical sequences occur before and after the loop |
|
|
cause of Huntington disease
|
an increase in the number of glutamine residues in huntingtin protein caused by bulging of triplet nu repeat CAG
|
|
|
sources of spontaneous/induced DNA damage (4)
|
1. reactive oxygen species
2. hydrolysis of glycosidic bonds (acid catalyzed) 3. hydrolysis/deamination of C/A 4. light induces formation of thymidine dimers |
|
|
direct damage repair
indirect damage repair |
direct - lesion reversed
indirect - enzymes cut out lesion |
|
|
example of reactive oxygen species damage
|
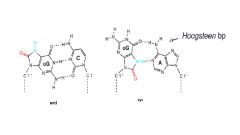
8-OxoG
- results in Hoogsteen bp with A |
|
|
example of depurination- hydrolysis of glycosidic bond
|
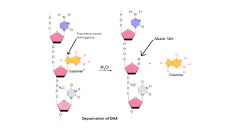
|
|
|
example of spontaneous deamination
|
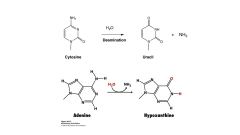
converts amino groups to keto groups by addition of water and elimination of ammonia
|
|
|
thymine dimers
- how is it repaired? |
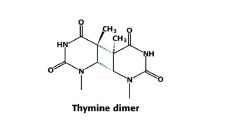
formed by reaction of UV light with two T's on same DNA strand
- reversible by enzyme DNA photolyase or repaired by uvrABC system |
|
|
what atoms in DNA bases are nucleotides?
|
N's & O's
|
|
|
example of DNA alkylation
|
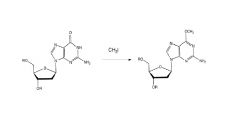
O6 and N7 are common alkylation sites; N3 is also reactive
|
|
|
Benzopyrene
|
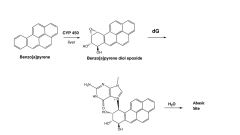
Cyp 450 converts to epoxide which can slip in between bases to bind and remove base -> abasic site
|
|
|
Aflotoxin B1
|
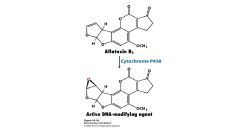
a natrual product synthesized by a mold that can be activated in the liver to a potent carcinogen
|
|
|
immediate mismatch repair during synthesis
|
edited directly by pol III (3' - 5'' exonuclease activity)
incorrect base pairing slows polymerase enoufh to allow the new DNA strand to enter the exonuclease site for hydrolysis - this activity increases accuracy of DNA synthesis by over 100-fold |
|
|
MutSLH repair
|
three part enzyme that repairs a mismatch
1. binds to site →activates endonuclease 2. nicks new DNA containing mismatch 3. exonuclease removes segment 4. DNA pol III fills in the gap 5. Ligase connects strands |
|
|
DNA methylation
|
allows identification of newly synthesized strands; repairs must occur before methylation
Dam methylase methylates new DNA strand |
|
|
uvrABC system
|
excision repair of thymidine dimers
1. excision of a 12 Nu fragmant by uvrABC exinuclease 2. DNA synthesis by DNA Pol I 3. joining by ligase |
|
|
excision repair of uracil and alkylated bases
|

1. recognition and hydrolysis by glycosidase
2. nicking DNA next to abasic sugar by endonuclease 3. repair by pol I 4. sealing by DNA ligase |
|
|
methylase
|
methelates a specific site in a DNA strand
- looks for a specific sequence then methelates one nucleotide |
|
|
AlkA
|
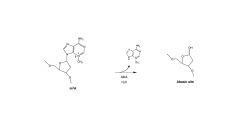
removes a mismatched nu to yield an abasic site for repair by pol I and ligase
|
|
|
8-oxoG repair
|
three enzymes: MutMYT
1. endonuclease cuts strand 2. exonuclease removes bases 3. ligase fills back in *** 8-oxoG can be more difficult to detect b/c it looks like a WC base pair*** |
|
|
DNA photolyase
|
direct repair of thyamine dimer- by SOME organisms
uses folate and flavin cofactors needs white light |
|
|
O6-MeGuanine Alkyltransferase
|
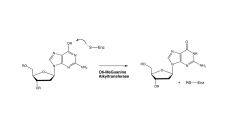
direct repair of alkylated guanin in DNA
- accelerates reaction but is not a true enzyme because it cannot turn over. Once the cysteine is alkylated, the enzyme is dead |
|
|
how does Ames test detect
|
a mutated strain that requires a specific medium for growth will proliferate around a disc of that medium placed on an agar plate
- used to detect genotoxic agents |
|
|
homologous recombination
|
the swapping of genetic segments between two DNA duplexes of similar sequence
- increases genetic diversity in sexual reproduction - can repair severely damaged DNA - process for deletion, swapping, or integration of foreign DNA segments into bacteria or other hosts * has been important for initial trials of gene therapy; normal human genes are exchanged for mutated copies in human cells |
|
|
crossover
|
homologous recombination
|
|
|
Rec A protein
|
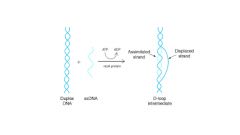
catalyzes strand exchange between homologous DNA molecules
- needs ATP |
|
|
Holliday junction
|
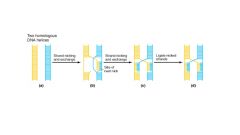
important intermediate of homologous recombination
- the process requires endonuclease to create nicks in phosphodiester backbone and recA enzyme to catalyze strand invasion and progression |
|
|
homologous recombination mechanism
|
1. endonuclease cuts
2. strand exchange 3. ligation 4. branch migration 5. resolution |

