![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
43 Cards in this Set
- Front
- Back
|
deoxythymidine
|
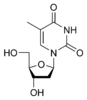
|
|
|
deoxyadenosine
|
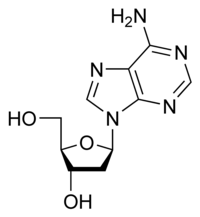
|
|
|
deoxyguanosine
|
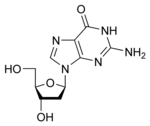
|
|
|
deoxycytidine
|
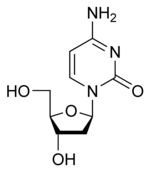
|
|
|
type of bond between ribose sugar and base
|
glycosidic bond
|
|
|
Base duplex formation mediated by:
|
Watson-crick H-bonding between complementary bases (basepairing)
|
|
|
draw a G-C base pairing
|
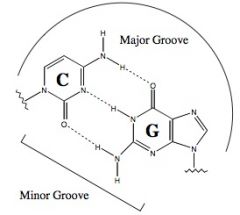
|
|
|
Draw an A-T base pairing
|
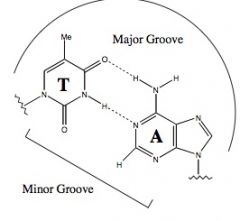
|
|
|
Which DNA surface (Major or minor groove) contains the most unique features for sequence specific DNA recognition?
|
Major groove
|
|
|
Major groove physical characteristics
|
-wider and deeper
-most DBD recognition here -Ave width: 11.6 A -Ave depth: 8.5 A |
|
|
Minor groove physical characteristics
|
-Ave width: 6.0 A
-Ave depth: 8.2 A |
|
|
which base pairing has more Propeller Twist?
|
more propeller twist in AT vs. GC base pairs
|
|
|
Some basepair parameters that affect the local conformation of the DNA
|
-Rise: Ave is 3.3 A
-Helical twist: Ave is 36 degrees -Propeller twist: ave is -11 degrees -Many more parameters exist |
|
|
Local conformation of DNA is _____ dependent (KEY POINT)
|
Sequence
|
|
|
How do DNA-binding domains exploit DNA conformation?
|
exploit sequence specific conformational flexibility/rigidity as well as local major/minor groove features to distinguish target sequence
-Example: AT regions usually have more compressed minor groove with hydration in minor groove |
|
|
Enthalpy in protein-DNA recognition
|
favorable: H-bonds, electrostatic interactions, non-polar interactions (van der waals)
Unfavorable: desolvation of polar groups and induced structural strain (DNA or Protein) |
|
|
Entropy in protein-DNA recognition
|
Favorable: Hydrophobic effect (release of ordered water form non-polar surfaces)
Unfavorable: reduced translational-rotational freedom, restricted vibrational motion, induced folding (protein) |
|
|
the energy of an electrostatic interaction is given by:
|

Coulomb's law
|
|
|
Hydrogen bond characteristics
|
-H-bonds in water typically have energies of 1-3 kcal/mol
-H-bond lengths range from 1.5-2.6 A -H-bonds are directional and polar |
|
|
van der Waals interactions
|
-Arise from asymmetric distribution of electronic charge around an atom
-Induces complementary dipole in nearby atom=attractive force -Interaction increases as distance decreases, until they are separated by van der Waals "contact" -Short distances=strong repulsion -VDW energies are small (.5-1 kcal/mol), but net effect over large molecules can be substantial |
|
|
EcoRI
|
-Cleaves GAATTC site 50,000X more efficiently than single base mismatch
-funtions as dimer: one monomer recognizes surrounding sequence, one monomer cleaves |
|
|
Phosphodiester cleavage by Type II restriction enzymes
|
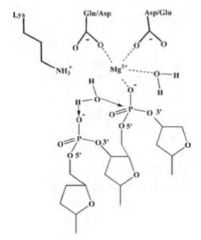
-SN2-type mechanism
-Water is attacking nucleophile after deprotonation by neighboring phosphate -Negative charge on attacked phosphate stabilized by Mg ion |
|
|
Equation representation of type II RE cleavage reaction
|
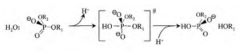
Pentavalent TS
product has inverted stereochemistry product= 5' monophosphate and 3' hydroxyl |
|
|
Method of studying the importance of specific contacts in a DBD
|
-synthetic chemistry
-Use base substitution to investigate the impact of certain natural base contacts on the free energy of DNA binding Ex: N6-methyl adenine, 7-deaza adenine, 2-deoxyuracil |
|
|
EcoRI base substitution study revealed:
|
binding is cooperative, not additive
|
|
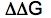
Major rule for binding
|
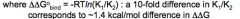
|
|
|
favorable energetic contributions of protein-DNA interaction largely offset by:
|
entropic penalties associated with complex formation
-Genomic DNA will act as a sink to decrease free soluble protein concentration |
|
|
specific rules for base-specific DNA recognition by Zinc finger proteins?
|
recognize adjacent DNA triplets
|
|
|
No rules for DNA recognition, but there are trends:
|
proteins take advantage of comlementarity between side chain donor/acceptor pairs and purine surfaces for recognition
-Arg likes N6 and O7 (usually G) -Gln/Asn likes N6 exocyclic amines (usually A) |
|
|
Affinity does not = specificity
|
Affinity is the difference in specificity between 2 different residues
|
|
|
The balance between enthalpy and entropy in driving protein-DNA association
|
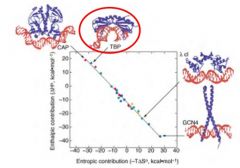
Either Enthalpic or Entropic contributions can drive association
|
|
|
TBP example
|
TBP is rigid, and bends DNA to bind
-Enthalpically unfavorable -Large hydrophobic surface is entropcically favorable, and this drives binding |
|
|
Some proteins bend DNA to bind; How is this achieved?
|
At lease 2 different mechanisms: wedges or phosphate neutralization
-Over short distances (<150bp) bends are disfavored due to rigidity from base stacking interactions and coulombic repulsion of phosphate backbone -TBP bends DNA by using "wedges": intercalating Phe side chains between neighboring base pairs -CAP uses noncovalent interactions and selective phosphate backbone neutralization to induce DNA curve |
|
|
The challenge of site-specific DNA recognition
|
Low protein concentrations (one individual protein) and high DNA concentration (per base pair) mean a ~4000 fold excess of non-specific DNA
|
|
|
Challenge of sit-specific DNA recognition: funtion
|

If the ratio of Ks/Kns is 10^3 and there are 4000-fold excess non-specific sites to specific sites, then 4-fold more protein will be bound to non-specific sites
-This problem increases with the size of the genome |
|
|
What problem does a large Ka^s present for a transcription factor?
|
Half life is lengthened, so response times are too slow.
|
|
|
How does a protein rapidly find it's preferred recognition sequence in the context of vast excess of non-specific sites?
|
Many proteins form a non-specific initial complex with DNA based on electrostatics.
-A layer of water + weak electrostatics allows the protein to slide rapidly along DNA and hop between DNA segments= combination of 1D and 3D search for its target sequence |
|
|
Biological systems use cooperative binding to enhance specificity and the rate of equilibrium:
|
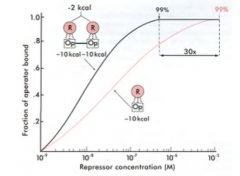
steeper slope=faster response
|
|
|
DNA damage recognition and repair
|
-Repair enzymes must identify modified bases in a "sea" of standard DNA: must discriminate between modified and standard bases
-hOGG1 recognizes 8-oxo-guanine when paired with C in duplexDNA (This modified base can pair equally well with C or A during replication=G-C to T-A transversion) |
|
|
Base excision repair mechanisms
|
1) hydrolysis to leave an Abasic site
2) Schiff base assisted hydrolysis and backbone cleavage |
|
|
One of the principle determinants exploited to recognize 8oxoG:
|
change of the N7 position from a H-bond acceptor to a H-bond donor
-Also recognizes "orphan" C if paired with A |
|
|
Sequence specific recognition of the major groove can also be mediated by:
|
A third strand of DNA (or RNA)
-binds major groove of DNA and recognizes the hoogsteen face of purine bases -Binding can be parallel (hoogsteen) or antiparallel (reverse hoogsteen)to the strand that is being recognized -This system provides a synthetic method for targeting desired DNA sequence |
|
|
purine-purine recognition using a third strand:
|
requires antiparallel arrangement of interacting strands (reverse hoogsteen)
|

