![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
29 Cards in this Set
- Front
- Back
|
2 techniques for in vitro selection of biopolymers
|
phage display
SELEX |
|
|
what is in vitro selection of biopolymers?
|
isolating a rare subpopulation from a large library (>10 members)
where all of the members are simultaneously assayed as a group (pool). The key feature of all of these techniques is a linkage between genotype & phenotype. |
|
|
phage display is a method for the selection of
|
proteins/peptides with a desired function
|
|
|
SELEX is a method for the selection of
|
functional nucleic acid motifs (both RNA and DNA)
|
|
|
General schematic overview of in vitro selection techniques (none in particular)
|
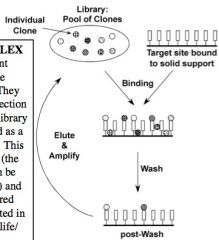
library members are screened in a pool, not individually
# of cycles depends on size of library and selection stringency ~bigger library=more cycles |
|
|
advantage of in vitro selection
|
assay conditions for the pool can be rigorously controlled
|
|
|
disadvantage of in vitro selection
|
desire clones cannot be isolated in a single step, unlike life/death genetic selection techniques
|
|
|
library construction for phage display and SELEX
|
libraries can be composed of anything from one randomized parent molecule to cDNA libraries
library must contain a link between genotype and phenotype library size for phage display can approach 10^10 library size for SELEX can approach 10^15 unique clones |
|
|
rounds of selection for phage display and SELEX
|
functional library members are separated from the pool based on affinity for a specific target or by colvalent modification when selecting for enzymatic activity
typically done by linking activity to the ability of clones to be retained on solid support altering conditions to be more stringent in subsequent rounds of selection narrows pool |
|
|
elution and amplification for phage display and SELEX
|
cycle is repeated multiple times until functional clones have been amplified sufficiently to comprise the majority of the pool
each completed cycle is termed a round |
|
|
Stringency
|
critical to ensure an enrichment of the desire clones during each round of selection, however in early rounds it is also critical to ensure that desired clones are not unintentionally discarded
stringency can be controlled at multiple points, including concentrations of library and target, buffer conditions, length of wash, ect increased stringency=decreased background of undesired clones |
|
|
stringency calculations
|
if 10 copies of each clone are present in the original library, then significantly more than 10% of positive clones must be retained on average
if only 10% of positive clones are retained, the proportion of positive sequences for which all members would be lost is (.9)^10 (35%) |
|
|
How does phage display technically work?
|
M13 filamentous bacteriophage
gene 8 encodes 1 major coat protein (3000 copies displayed along entire phage genes 3,6,7,9 encode 4 minor coat proteins (5-6 copies displayed on one end or the other) fusion proteins usually made on gene 8 or 3, depending on desired level of display ~using gene 8 can create a pseudo-cooperative binding situation ~often use gene 8 to identify an initial lead, followed by optimization using gene 3 or other methods |
|
|
generating a phage library
|
use a "phagemid"
~plasmid containing the fusion gene and phage f1 origin ~each bacteria contains only one member of the library= no scrambling between genotype and phenotype |
|
|
Schematic view of phage display technique
|
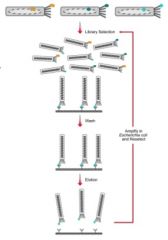
|
|
|
phage display selection
|
sometimes a competitor is used to increase stringency
some may bind to solid support, so change solid support between rounds, or have a specific way to release phage + target from support (like cleave target) stringency can also be adjusted by changing concentration of target/phage particles |
|
|
phage display wash
|
typically one of the key stages that determines stringency of selection
(#/length of washes) |
|
|
phage display elution
|
removed from solid support via acid, high salt, proteolysis, ect
|
|
|
phage display amplification
|
eluted phage allowed to infect e. coli
e.coli are amplified and infected with helper phage to generate a new pool of phage to use in the next round of selection |
|
|
some practical uses of phage display
|
-selection of protein(peptide)-protein, protein(peptide)-DNA/RNA interactions
-mapping of protein-protein interaction domains -selection of small molecule binding domains -cDNA library searches for interacting partners -selection of novel substrate specificity for an enzyme (catalytic antibodies) |
|
|
protein engineering example: selection of dimeric zinc finger protein
|
dimerization domain and DNA binding domains combined using varied "linker" lengths
Amino acids were randomized, but cystein was not used to avoid disulfide bond false positives DNA with desired target sequence was linked to solid support, and nonspecific DNA in solution eliminates false positives from nonspecific binders |
|
|
Methods of phage display library construction
|
-cassette mutagenesis
-kunkel mutagenesis -mutagenic PCR -DNA shuffling |
|
|
DNA shuffling
|
power technique for "evolving" genes during selection
-incorporates a stage of recombination to allow mutations that improve fitness to be dispersed through the library population |
|
|
limitations of m13-based phage display
|
successful display of fusion protein requires that the fusion protein be successfully exported to the periplasmic space
~using signal recognition particle (SRP) translocation pathway increases the types of proteins that can be displayed growing coat protein must be extruded through pores in outer membrane -proteins <30kD display efficiently -proteins approaching 100kD are displayed inefficiently or not at all -displayed proteins must fold without chaperones in the periplasmic space |
|
|
limitations of m13 phage display library construction
|
fusions must be made n-terminal to the phage coat protein (c-term is assembled into the phage partical coat)
-limits usefulness of m13 for displaying cDNA libraries, which inherently contain stop codons ~overcome problem by using dimerization domain to link the library protein to the coat protien |
|
|
what does SELEX stand for?
|
systematic evolution of ligands through exponential enrichment
|
|
|
SELEX advantages
|
fully in vitro system (no transformations nessesary)
~means very large libraries can be used |
|
|
SELEX disadvantages
|
# of functional groups is much more limited than for proteins
~bypass problem by using cofactors |
|
|
SELEX steps
|
-PCR synthetic DNA pool to create dsDNA pool
-in vitro transcribe (T7 RNA POL) to RNA -use binding selection to enrich for certain RNA -Reverse transcribe to cDNA -PCR to re-attach T7 promoter and regenerate dsDNA pool -repeat selections until satisfied -clone final dsDNA pool to isolate aptamers |

