![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
57 Cards in this Set
- Front
- Back
|
What's the biological importance of recombination and transposition? |
Transposition gives rise to spontaneous mutations. Viral infections and acquired immunity come from recombination.
Site specific recombination is used to insert and excise viral genetic material as well as alter gene expression |
|
|
What are the 3 types of CSSR? |
Conserved site specific recombination
1. insertion, 2. deletion 3. inversion
|
|
|
How does recombination work? |
At recombination sites, recombinase recognizes sequences in the crossover region (the area where the base pairing is identical) It then binds to the sequence and recombines
|
|
|
Chemistry of recombinase via serine recombinase (similar to topoisomerase) |
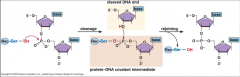
|
|
|
What is different about tyrosine recombinase vs serine recombinase? |
One of the key differences between the serine and tyrosine recombinases is that tyrosine recombinase reaction a branched DNA intermediate or "Holliday Junction" forms after the first strand exchange, whereas Holliday junctions never form during a serine recombinase reaction. Cre. recombinase and the lambda integrate are examples of tyrosine recombinase. |
|
|
Tyrosine recombinase Mechanism 1 (one crossover at a time) |
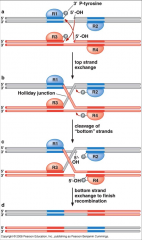
|
|
|
Tyr recombinase mech 2
|
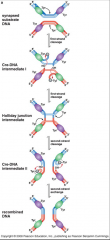
|
|
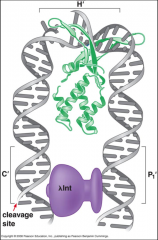
What is this?
|
The green is the IHF (integration host factor) and induces a bend in the DNA so that lambda integrate can do its work cutting and inserting the new genetic material. The weak core sites are closer to each other, which in turn leads to the catalyzed recombination |
|
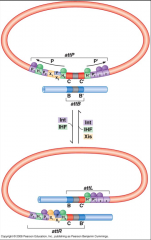
|
Architectural proteins ensure that excision and incision are occurring at the correct time. You have to bind the recognition factors before recombination can occur
|
|
|
Why is CSSR conservative? |
No need fore external energy. recombinase does the breaking and the resealing |
|
|
How many subunits of serine recombinase are needed for recombination? |
4 |
|
|
What does the structure of serine recombinase suggest about the mechanism? |
There are 4 units in a complex. The recombinase dimers have a section that is hydrophobic and allows for rotation to occur. Also some regions of +/- charge help stabilize the DNA in the initial or 180 degree state. |
|
|
Cre recombinase |
Cre is an enzyme encoded by phage P1 which functions to circularize the linear phage genome during infection. It acts on recombination sites on the DNA called lox sites. It requires 4 subunits of Cre with each molecule bound to one binding site on the substrate. It uses a holliday junction intermediate. |
|
|
Cre recombinase - medical application |
some site-specific recombination systems are so simple that they are widely used as tools in experimental genetics. For example, genetisits can insert recombination sites into the gene of interest. This won't affect gene function unless the recombinase is present. (sometimes the function is needed in the early stages of life) |
|
|
What is an architectural proteins? |
An accessory protein required for certain recombinations. They bind specific DNA sequences and bend the DNA. They organize DNA into a specific shape and thereby stimulate recombination. They can also control the direction of a recombination reaction. |
|
|
What is the importance of accessory proteins? |
They ensure that the DNA integration and excision occur at the right time in the phage life cycle. |
|
|
Salmonella Hin Recombinase |
inverts a segment of bacterial chromosome to allow expression of two alternative sets of genes. In Hin recombinase, it is used to evade a bacterial host immune system |
|
|
hixL and hixR in Salmonella Hin recombinase |
specific recombination sites (L for left and R for right) They are inverted with respect to each other. If they are ON then H2 flagellin is expressed because the promoter fljB (H2 promoter) and fljA (H1 repressor) are expressed. In the OFF state, then neither promoter is active and H1 flagellin is expressed |
|
|
What does Fis do in relation to Hin recombinase |
It is an enhancer that increases the rate of recombination 1000 fold. It's like IHF in the sense that it is a site-specific DNA-bending protein (unlike IHF, it is not essential for recombination). It is greatly facilitated by negative supercoiling which stabilizes association of distant DNA sites |
|
|
What is a resolvase? |
They resolve dimers and multimers into monomers (for circular DNA) |
|
|
Xer Recombinase is a member of which recombinase family? |
Tyrosine |
|
|
Xer |
A heterotetramer, containing two subunits of XerC and two subunits of XerD. Both are tyrosine recombinase but they recognize different DNA sequences. |
|
|
How do cells make sure Xer-mediated recombo at dif sites will convert the chromosome into monomers? |
Xer recombinase works with cell division protein FtsK. If FtsK is inactive, then only the XerC is active and a holliday junction intermediate is formed but the reversal occurs because XerD is inactive and can't complete the recombination |
|
|
What is FtsK |
an ATPase that track along DNA. It helps regulate XerC/XerD and moves DNA from the center of cell during division. |
|
|
What is transposition? |
A specific from of genetic recombination that moves genetic material from one DNA site to another. These mobile elements are called transposable elements or transposons. |
|
|
How is transposition different from site specific recombination? |
Not specific sequence. The enzymes that catalyzes transposition are often encoded in the transpose itself. Often disrupt gene function and are the most common source of new mutation. |
|
|
What are the two modes of transposition? |

|
|
|
What are the 3 types of transposable elements? |
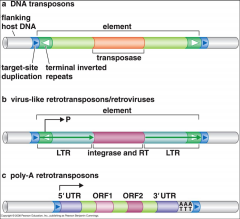
|
|
|
What is reverse transcriptase? |
A special type of DNA polymerase that can use RNA template to synthesize DNA. |
|
|
What is the difference between retroviruses and retrotransposons? |
The genome of the retrovirus is packaged in a viral particle and escapes the host cell to infect other cells. The retrotransposons can only move to new DNA sites within the cell, they can never leave. |
|
|
Explain the initiation of the cut-and-paste transposition |
Initiation: binds to the terminal inverted repeats at the end of the transposon. The two ends are brought together by transposases to form a stable synaptic complex or transpososome. |
|
|
Explain the excision of the cut-and-paste transposition |
The transposases will cut the DNA so that there are free 3'OH ends and they will cut one strand at each end of the transposon. The second strand is cut with a different mechanism |
|
|
Explain incision of the cut-and-paste transposition |
Once the transposon is free, the ends attack the DRNA phosphodiester bonds at the site of the insertion. This segment of DNA is called the target DNA. The transposome insures that the DNA strands attack the same target sites. |
|
|
What are the 3 mechanisms for cleaving the non transferred strand |
1. An enzyme other than transposes is used (like TnsA which acts like a restriction endonuclease) 2. The transposases catalyzes the attack of one DNA strand on the opposite strand forming a DNA hairpin intermediate. 3. The hermes transposon uses a second mechanism of second strand cleavage by hairpin formation. The cleavage in this case is of the top strand (non transferred strand) first and then the hairpins generate at the original insertion site rather than the transposon end |
|
|
Hermes transposon is similar to... |
antibody VDJ recombination |
|
|
Why don't more transposons follow replicative transposition? |
Often causes chromosome inversions and deletions causing selective disadvantage for the host. |
|
|
Describe retroviral integration/virus-like retro-transposon transposition |
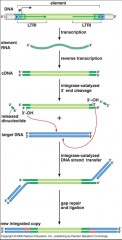
|
|
|
What are DDE-motif transposes/integrase proteins? |
They have a common catalytic domain which contains three invariant amino acids. 2 aspartates and 1 glutamate. These AAs form part of the active site and coordinate divalent metal ions. The unique thing is that the same active site is for cleavage and transfer. |
|
|
Tn5 transposase DNA complex |
The transposase subunit that is bound to the recombinase recognition sequences on one of these DNA fragments (i.e., on one transposon end) donates the catalytic domain that promotes the DNA cleavage and DNA strand transfer reactions on the other end of the transposon. Because of this subunit organization, the transposase will be properly positioned for recombination only when two subunits and a pair of DNA ends are present together in the complex. |
|
|
What is the poly-A retrotransposon mechanism? |
Target-site primed reverse transcription. 1. Transcription of the DNA an integrated element by cellular RNA Pol. Promoter is embedded in the 5'UTR 2. RNA is translated to ORF1 and ORF2 which stick around RNA 3. Protein/RNA complex then re-enters the nucleus and associates with the cellular DNA. 4.The complex binds to a T-rich site in the target DNA 5. The proteins initates cleavage in the target DNA leaving a 3'OH for binding. 6. Reverse transcriptase + line element |
|
|
Orf2 |
ORF2 has a DNA endonuclease activity and a reverse transcriptase activity.
|
|
|
What do LINE and SINE stand for |
Long interspersed nuclear element Short interspersed nuclear element |
|
|
SINE vs normal mRNA that's inserted |
SINE doesn't have any encoding sequences |
|
|
Tn10 what is it? |
A compact element of 9kb and encodes a gene for its own transposes and gene imparting resistance to tetracycline |
|
|
How does Tn10 work? |
follows the cut/paste mechanism. The eft transposes is defective |
|
|
How is Tn10 controlled? |
The overlapping promoter region has an IN (left) and an OUT (right). The P(out) will promote the expression of an antisense RNA. The antisense is longer lived than the mRNA of the P(IN). In cells with high number of Tn10, the RNA:RNA pairing is frequent (antisense and sense) and the number of copies are reduced |
|
|
Mu phage transposon |
Is 40kb carrying 35 genes; only MuA and MuB are need for transposition; it is very active (100 transpositions per hour) |
|
|
How does Mu phage transposon avoid transposing itself? |
Transposition target immunity - MuB is an atpase; however in the presense of MuA, it hydrolyzes the ATP of MuB so that it doesn't bind to DNA (inactve) |
|
|
Ty elements - what are they? |
transposons in yeast that are virus-like retrotransposons. They're even packaged into virus-like particles. They tend to transpose very specific regions. |
|
|
Where do Ty1-4 integrate and why? |
Upstream of the tRNA promoters because their integrate interacts with the Pol III polymerase responsible for tRNA transcription |
|
|
Where does Ty5 integrate? Why? |
In the silent genetic regions like the telomeres and the silent copies of the mating loci. Binds to DNA-silencing protein Sir4. |
|
|
Why do Ty elements exhibit regional target site preference? |
It enables the transposons to persist in the host by focusing most of their insertions away from important regions of the genome that are involved directly in coding for proteins. |
|
|
What is V(D)J recombination |
The principle method of generating antibodies and T-cell receptors |
|
|
How many combos of antibodies are there? |
2 constant regions for light chains. heavy + light chain combos = 5 million combos! |
|
|
How does VDJ recombo work? |
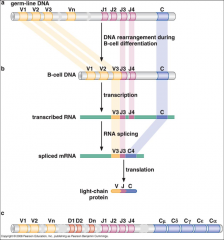
|
|
|
Recombination sequences (recombination signal sequences) |
Recombo always occurs between two recombo signal sequences. RAG1 and RAG2 are responsible for recognizing and cleaving the recombination signal sequences |
|
|
V(D)J segments are brought together by non homologous end joining. |
Cleavage occurs in a mechanism very similar to transposon excision. The recombinase catalyze single-strand cleavage at the ends of the signal sequences leaving free 3'OH. They initiate attacks on the other ends forming hairpins. The two hair pins are hydrolyzed and combined. This under goes further recombination and the strand with the two recombination signal sequences are formed into a signal joint and discard |

