![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
23 Cards in this Set
- Front
- Back
|
What is the lac operon? |
Operon: cluster of structural genes that are coordinately regulated by a single promotor region (During transcription, all structural genes aretranscribed onto one polycistronic mRNA molecule. During translation, each gene is translatedindependently, to produce several gene products) The lac operon has a cluster of 3 genes (lacZ, lacY, andlacA) that encode the 3 enzymes (β-Galactosidase,permease, and transacetylase) that E.coli uses to digest thesugar lactose. |
|
|
What happens to the lac operon when glucose is present/absent? |
When E.coli lacks glucose (its preferred carbon source) butlactose is present, the lac operon is expressed, and all 3genes are expressed together, in roughly equal amounts. When there is glucose present (or no lactose present), theentire lac operon is not expressed, and none of theseunnecessary enzymes are produced. |
|
|
Explain the non-induced lac operon process. |
- When there is no lactose, the lac operon should not be expressed because the 3 gene products are not needed. - expression of the lac operon is blocked by the binding of a specific lac repressor protein to a specific binding sequence in the lac operon called the operator (operator DNA sequence found between the promoter and the transcription start point, so that binding a repressor there blocks RNA polymerase from binding there, blocking initiation of transcription) |
|
|
What is the lac I gene? |
It encodes the repressor protein and is not part of the lac operon itself. Lac I is expressed separately, so that there is always enough lac repressor protein in the cell, to regulate the lac operon. |
|
|
Explain the induced lac operon process. |
- When lactose is present (and glucose absent), the lacoperon should be expressed, since its 3 genes’ enzymes arenow needed! - Lactose (the inducer) binds to an allosteric site on therepressor protein, changing its shape so that it can’t bindstrongly to the operator sequence of the lac operon. - As a result, the repressor protein detaches from theoperator sequence, and RNA polymerase can now bind tothe promoter sequence for the lac operon, and begintranscribing its coding sequences. |
|
|
How do you produce a merodiploid strain? |
Merodiploid strain: bacterial strain with two copies of thatgene/operon. - A crossover between the ISs on either side of the integrated Fplasmid would excise the F plasmid (turning Hfr cell to F+ cell). But crossovers between more distant ISs can excise someadjacent chromosomal genes, too, to make an F’-lac plasmid. - If we use conjugation to add this F’-lac plasmid to an F- cell thatalready has a lac operon, we would produce a merodiploid(i.e. a partial diploid) that has two copies of the lac operon: - one in the bacterial chromosome, and - one extra copy in the F’-lac plasmid |
|
|
Which alleles are trans-acting and which are cis-acting? |
Alleles that make proteins tend to be trans-acting,since the proteins can travel to both operons, whereas DNAsequences (like the operator) must stay put, and be cis-acting. |
|
|
Describe the lac operon mutations with merodiploids. |
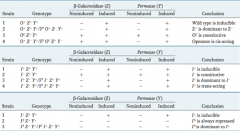
|
|
|
What are the steps to predict gene expression? |
POIG (Ex I- P- Oc Z+ Y+/F’ I+ P+ O+ Z- Y-) - Because if there’s a P-, there will be no expression of theadjacent structural genes (so you can cross them off). If there’s a P+, go to step 2. - Because if there’s an Oc, the adjacent structural genes will beconstitutively expressed (regardless of I alleles or induction). If there’s an O+, go to step 3. - If there is at least one Is allele, the super-repressor it encodeswill permanently repress all O+ operons (since it acts in trans). If at least one I+ (and no Is), then normal gene expression for allO+ operons (when induced by lactose). If only I- alleles available, then no repressor present, and so all P+ operons are constitutivelyexpressed (since no repression) 4) Look at the structural genes, to see if they can make enzymes (+). In this example, the second operon (with P+ and O+) doesn’t haveany wildtype structural genes, so no enzymes are made.
|
|
|
What effect should the absence of glucose have on lac operons, when lactose is present? |
Lac operon expression should be promoted with an activator. When there’s not enough glucose, cells need more lac enzymesto use the available lactose as a carbon/energy source |
|
|
When glucose is abundant, why is there much less expression of thelac operon? |
Ultimate = digesting lactose takes more time/energy than glucose. Proximate = the activator protein isn’t bound to the lac operator. |
|
|
What is positive/negative regulation in |
Positive regulation: Even when transcription is not A repressor protein binds to theblocked by a repressor, very little operator sequence (between P transcription happens unless anactivator binds to the activator-binding site (beside promoter).The activator helps RNApolymerase bind more optimallyto the promoter (to increase transcription) Negative regulation: a repressor protein binds to the operator sequence (between P transcription happens unless anactivator binds to the activator-binding site (beside promoter).The activator helps RNApolymerase bind more optimallyto the promoter (to increase transcription). and the coding sequences) toblock RNA polymerase frombinding (blocks transcription). |
|
|
What are activators and repressors? |
They are both regulatory proteins. Therefore, they can both regulate expression of genes/operons bybinding to specific DNA sequences (with their DNA-binding sites). They can also “sense” environmental conditions by binding to specific effector molecules with their allosteric sites. Binding toeffector molecules changes regulatory proteins’ DNA-binding ability. |
|
|
What do CAP and cAMP do? |
CAP = lac activator protein cAMP = the effector for CAP When there isn't enough glucose, ATP gets converted to cAMP. cAMP binds to CAP and the resulting cAMP-CAP complex binds near the lac promoter and improves binding of RNA polymerase to the promoter. This increases expression of the lac operon. When there is lots of glucose, no cAMP accumulation, & nocAMP-CAP complexes form. CAP does not bind to DNA on itsown. Without this activator, binding of RNA polymerase to thepromoter is inefficient, and so there is very little lac operonexpression. |
|
|
What are the similarities and differences between the ara operon and the lac operon? |
They both have 3 structural genes but ara has B, A, D and lac has X, Y, A. These structural genes encode enzymes for catabolizing sugars but the sugars they catabolize are different: arabinose vs lactose. For ara the operator is upstream to the promotor, for lac it's downstream. There is a gene outside of the operon for the regulatory protein but the C gene makes the araC protein and the I gene makes the repressor protein. |
|
|
How is negative regulation different in ara than in lac operon? |
Lac: when lactose is absent the lac repressor binds to lacO, which blocks transcription by preventing RNA polymerase binging. Ara: when arabinose is absent the araC protein also binds binds to araO to block transcription but it also binds to araI (initiator region). This bends DNA into a loop, which blocks binding of both RNA polymerase and the CAP-cAMP complex. |
|
|
How is positive regulation different in ara than in lac operon? |
Lac: when lactose is present and glucose is absent, lactose binds to the repressor and detaches is from lacO. Concentration of cAMP increases, so CAP-cAMP binds near the promotor to greatly increase expression of the structural genes. Ara: when arabinose is present and glucose is absent, arabinose binds to araC protein and detaches is from araO. Concentration of cAMP increases, so CAP-cAMP binds near the promoter to greatly increase expression of the structural genes. ** both use CAP and cAMP for positive regulation but each operon is still repressed until its target sugar (arabinose / lactose) is thereto remove the regulatory protein (araC protein / lac repressor)from its operon. |
|
|
Why does the araC protein remain attached to the araI when arabinose is present? |
AraC protein has a second function when arabinose ispresent: The arabinose-AraC protein complex binds to araI tofacilitate binding of RNA polymerase and to initiate moretranscription. So this dual-function araC protein exhibits dualcontrol (both negative and positive) since it acts as both arepressor (when arabinose is absent) and an activator (whenarabinose is present) |
|
|
What is attenuation? |
Transcription is terminated early (at theattenuator), when there is lots of, for example, tryptophan available. Transcription review: Transcription ends when RNA polymerase reaches the templatestrand’s termination sequence ( = a sequence of ~40 bps thatstops transcription). This termination sequence has two important regions near its end: a GC-rich region, followed by a string of “A” nucleotides. The GC-rich region has two-fold symmetry, so that the RNA transcript can fold over to base-pair with itself, forming thisstable, GC-rich RNA hair-pin stem loop. |
|
|
Describe the intrinsic mechanism of the termination of transcription. |
1) RNA polymerase starts transcribing the termination sequence. 2) Once transcribed onto RNA, the GC-rich region of the RNA folds and base-pairs with itself to form the hairpin stem loop. 3) RNA polymerase then adds a series of U RNA nucleotides, to match the string of A nucleotides in the termination sequence. 4) This string of A-U pairs are unstable, so RNA polymerase backtracks to seek a more stable G-C pair before continuing. 5) The hairpin loop blocks access to any G-C pairs, so RNA polymerase “gives up” and releases theRNA and DNA. |
|
|
How does this termination mechanism relate to trp operons? |
There is a long leader sequence that’s transcribed onto RNAbefore any structural genes. That leader includes an earlytermination sequence called an attenuator (when it’s transcribed,the mRNA forms a loop just before some unstable U-A base pairs). When high [trp], the loop persists, and transcription ends early.When low [trp], the loop is opened, and the ribosome canbacktrack to find a stable G-C pair before moving forward andtranscribing the rest of the operon. |
|
|
How does the ribosome disrupt the loop when [trp] is low? |
- When the leader sequence is transcribed onto RNA, there are4 regions that can base-pair with one of their adjacent regions. - When regions 1 and 2 are base-paired, regions 3 and 4 canbase-pair to form the loop that ends transcription early. - But when [trp] is low, the ribosome gets stuck in region 1 (b/c itcan’t translate 2 trp codons). With region 1 blocked, region 2binds to region 3 instead. With the loop between regions 3 + 4opened up, RNA polymerase can backtrack into region 4 to finda G-C pair, and then continue transcribing the rest of the operon. |
|
|
How does the ribosome disrupt the loop when [trp] is high? |
But when [trp] is high, the ribosome doesn’t get stuck in themiddle of region 1... instead, it pauses at a stop codon at the endof region 1, and blocks region 2 from binding to region 3. -As a result, regions 3+4 stay bound together as a loop, and RNApolymerase must detach, thus terminating transcription early. |

