![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
63 Cards in this Set
- Front
- Back
|
point mutations |
cause folding issues with the resulting proteins |
|
|
deletion mapping |
1. allow us to determine the location of point mutations 2. use known deletions to map the location of a point mutation within the same gene |
|
|
what strains of E. coli will we use? |
F+ and F= |
|
|
F+ and F- strains |
1. E. coli strains 2. conjucage and pass genetic information from donor F+ strain to recipient F- strain 3. will conjugate and transfer the deleted portion of the Z gene from donor cell's plasmid DNA to the recipient cell |
|
|
F+ strain |
1. donor strain 2. have Fertility Factor 3. have Z gene in plasmid DNA with some part of the Z gene sequence deleted 4.can have multiple F+ strains with different portions of the Z gene (some will rescue the Z gene after recombination, others wont) |
|
|
donor strain |
F+ strain |
|
|
F- strain |
1. recipient strain 2. have point mutations in Z gene 3. enzyme won't fold properly 4. organism can't utilize lactose as a C/E source |
|
|
recipient strain |
F- strain |
|
|
plasmid DNA |
contains the Fertility Factor |
|
|
Fertility Factor |
1. allows conjugation to occur 2. transfer the plasmid to the recipient cell |
|
|
conjugation |
transfer of genetic information from donor to recipient |
|
|
B- galactosidase gene |
1. allows organism to use the alternative carbon/energy (C/E) source lactose 2. Lactose ---B galactoseidase---> glucose + galactose |
|
|
lactose |
1. alternative C/E source 2. Lactose ---B galactoseidase---> glucose + galactose 3. lactose is broken down into glucose (proper substrate) |
|
|
glucose |
1. broken down from lactose 2. proper substrate for the enzyme hexokinase (for glycolysis) |
|
|
hexokinase |
start of glycolysis |
|
|
recombination |
1. can occur between the donor's plasmid DNA and the recipients genomic DNA 2. can swap out point mutation using the DNA offered by the donor's plasmid DNA --> lactose utilization
|
|
|
recipient gDNA |
1. has a point mutation that caused Z gene product to fold improperly 2. can't utilize lactose |
|
|
donor plasmid DNA |
if it contains normal portion of DNA that has point mutation in recipient, recombination can swap out point mutation using DNA offered by donor's plasmid DNA
|
|
|
post recombination: recipient |
1. recipients genome no longer has point mutation 2. has WT Z gene 3. recipient can utilize lactose as C/E source 4. can now grow on media that has lactose instead of glucose |
|
|
F+ multiple strains |
1. some may rescue Z gene during F- recombination, others wont 2. have different portions of Z gene deleted in its plasmid DNA 3. must first know what portion of the Z gene has been delted from the Z gene located on the Donor's plasmid DNA |
|
|
CSH |
1. cold spring harbor 2. designation for the strain of F+ E. coli distributed by CSH |
|

|
before recombination: 1. point mutation on recipient gDNA 2. recpient can't utilize lactose as C/E source |
|
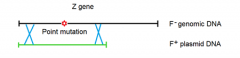
|
1. recombination occurring 2. transfer of point mutation |
|

|
1. post recombination 2. recipient now has functional WT Z gene (can utilize lactose as C/E source) 3. point mutation now in donor plasmid DNA |
|
|
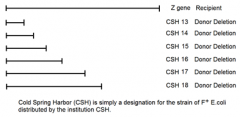
1. shows the recipient's Z gene which will have the point mutation somewhere in its sequence 2. shows what portion of Z gene has been deleted from the sequence located in the donor's plasmid DNA more important to think about what DNA is present on the Donor plasmid DNA so recombination can occur and restore WT Z gene |
|
|
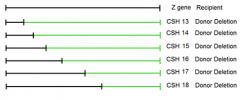
1. have deletions of the Z gene which have a known portion of the gene delted 2. if we mate a F- to each of the deletion F+ strains, some deletions will recover the WT gene, others won't depending on point mutation location 3. analyzing those that do recover and don't recover the ability to utilize lactose as a C/E source will determine the location of the point mutation |
|
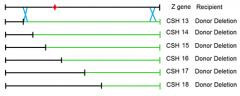
would mating of the CSH 13 strain to the recipient point mutant recover the WT Z gene? |
Yes; it would restore the ability of the organism to utilize Lactose as a C/E source |
|
|
would mating of the CSH 13 strain to the recipient point mutant recover the WT Z gene? |
can show the location of the point mutation of the recipients Z gene |
|
|
what do we use to identify the location of the point mutants? |
1. known deletions of the Z gene 2. conjugation 3. recombination 4. plating on Lactose media |
|

|
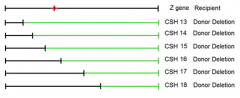
data seen after plating on Lactose C/E source media when point mutation is at this location
|
|
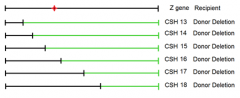
what growth data would we see after plating this bacteria with the known point mutation location on a Lactose C/E source media? |

|
|
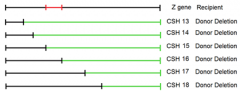
|
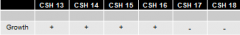
1. the best we could do with these deletion mutants is to narrow it down to the region highlighted in red 2. more deletions of different sizes could narrow it down further |
|
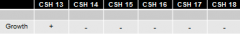
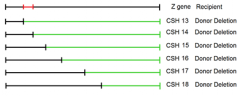
identify the region with the Z gene which contains this point mutation:
|
|
|
identify the region with the Z gene which contains this point mutation: |
|
|
|
if the point mutation lies within the deleted region ______________________ |
you will not recover enzyme activity |
|
|
if the point mutation lies outside the deleted region ______________________ |
you will recover enzyme activity |
|
|
CSH strains 13-18 |
1. conveniently deleted subsequently more and more of the gene from one end of hte gene 2. won't always be the case 3. might have to map locations of point mutants using randomly deleted amounts of the gene under observation |
|
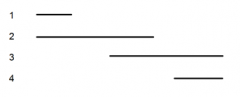
|
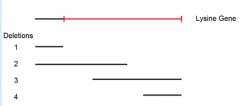
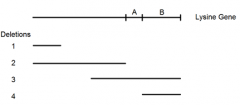
shows the deleted regions (1-4) of an enzyme used in lysine synthesis |
|
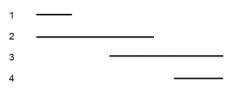
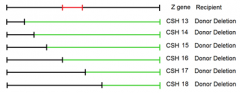
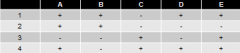
5 lysine point mutations (A-E) are mated to these 4 deletion mutants an dtested for ability to synthesize own lysine |
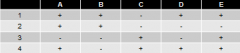
"+" = growth on - Lys paltes |
|
identify possible locations of each of the five point mutations. (First locate point mutant A) |
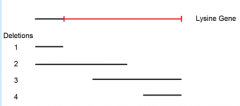
|
|
|
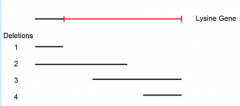
point mutant A mated to deletion mutant 1 yields these possibilities for the location of point mutant A |
|
|
point mutant A mated to deletion mutant 1 yields these possibilities for the location of point mutant A |
|
|
|
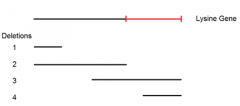
point mutant A mated to deletion mutant 2 yields these possibilities and narrows the location of point mutant A |
|
|
point mutant A mated to deletion mutant 2 yields these possibilities and narrows the location of point mutant A |

|
|
|
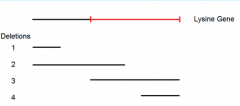
point mutant A mated to deletion 3 yields these possibilities but doesn't advance our knowledge |
|
|
point mutant A mated to deletion 3 yields these possibilities but doesn't advance our knowledge |
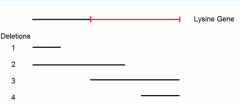
|
|
|
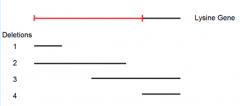
Point mutant A mated to deletion mutant 4 yields these possibilities giving us more knowledge of its location |
|
|
Point mutant A mated to deletion mutant 4 yields these possibilities giving us more knowledge of its location |
|
|
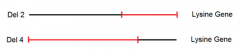
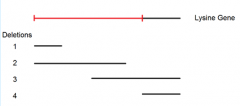
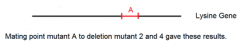
combining data from mating point mutant A to both deletion mutants 1 and 4 yields: |
|
|
|
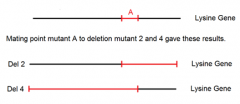
how to map the possible locations of point mutation A |
1. using matings between point mutant A and four deletion mutants 2. can now do the same for point mutants B-E using the same data table and thought process |
|
|
|
|
|
DIY deletion mapping: mutant B
|
|
|
|
DIY deletion mapping: mutant C |
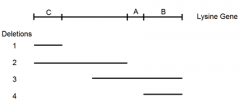
|
|
|
DIY deletion mapping: mutant D |

|
|
|
DIY deletion mapping: mutant E |
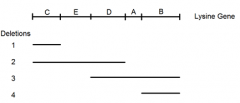
|
|
|
where do point mutants lie? |
recipient E. coli gDNA |
|
|
What are we passing from donor to recipient? |
passed donor's plasmid DNA through to our recipient's gDNA that have deleted portions of the Z gene |
|
|
does donor gDNA contain Z gene? is it functional? |
1. yes 2. it is functional 3. can break down lactose into glucose for utilization in glycolysis |
|
|
How do we know the resulting growth on lactose plates is the result of recombination and not from the growth of donor E.coli (since it has a functional gDNA Z gene)? |
1. donor strain of E. coli is sensitive to streptomycin 2. lactose plates contain streptomycin 3. therefore donor strains will not grow |
|
|
growth on plates |
represent streptomycin resistant recipient cells that have recombined with donor plasmid DNA and restored function of Z gene |
|
|
LAB: what are the 4 different point mutants we will identify for possible locations? |
CSH 1, CSH 4, CSH 6, CSH 9 |
|
|
LAB: what are the deletion mutants we will be mating the point mutants with? |
CSH 13, CSH 14, CSH 15, CSH 16, CSH 17, CSH 18 |
|
|
Use the chart previously shown regarding the deletions mutants CSH 13-18, determine possible locations of pt mutations CSH 1-4 |
* |

