![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
35 Cards in this Set
- Front
- Back
|
proteins are composed of?
|
amino acids
- individual amino acids have different properties, therefore amino acids give these different properties to the proteins that contain them. |
|
|
come characteristics of amino acids:
|
- some are polar or non polar
- some kink the protein - some are negative or positively charged - some can hydrogen bond - some can ionically bond to other amino acids - some can covalently bond with other amino acids |
|
|
Primary protein structure:
|

the linear sequence of amino acids in a protein.
1. linked by peptide bonds (condensation synthesis of amine and carboxyl groups of adjacent amino acids) 2. have linear directionality with an amine end and a carboxyl end. 3. protein size: usually between 50 - 2000 amino acids long |
|
|
Secondary protein structure consist of what two common types:
|
1. alpha helix
2. beta sheets |
|
|
Alpha helix:
|
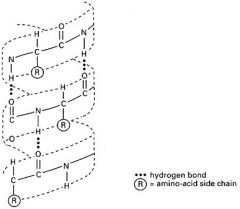
- hydrogen bond interactions between closely placed amino acids (ketone O and amine H)
- regular and repeating: throws strand into a uniform helix - precludes certain amino acids "Kink" the strand (ex. proline) - make that region of the protein more "linear" |
|
|
Beta pleated sheet:
|
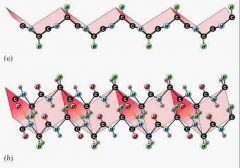
- caused by hydrogen bonds between amino acids in either two different proteins or two different regions of the same protein
- often beta sheet is regular and repeating as well: throws protein strand into a lampshade and H bonds ties the 2 beta sheet together |
|
|
Tertiary Protein structure:
|
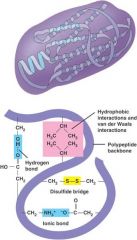
very important in determining the 3D structure of the protein
|
|
|
What causes the 3D structure in a tertiary protein?
|
- hydrophobic/ hydrophilic interactions between far apart amino acids
- polar/ nonpolar interactions between far apart amino acids - ionic bonds interactions between far apart ionic amino acids - covalent bonds (cys-cys) between far apart sulfur containing amino acids (disulfide bridge) ** occurs because it take less energy for reactions to occur rather than keeping it apart** |
|
|
What effects of tertiary structure have on protein?
|
1. the tertiary interactions bend protein strand on itself
2. gives the overall 3D shape, which mostly will be the lowest energy confirmation. 3. makes loops and has the effect of making the protein globular and possibly forming pockets in the proteins (active sites/ allosteric sites in enzymes) 4. shape is a balance of torque that the various interactions induce and the stress of bending the molecule. |
|
|
Quaternary protein structure:
|
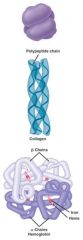
proteins made up of multiple protein subunits.
subunits are translated separately and then assembled into the holoprotein (in golgi or sER) |
|
|
Prosthetic Groups:
|
- are ions, sugar, metals, lipids tha tare covalently bonded to the protein. Often these prosthetic groups are important in the function of the protein
- Fe, Cu, Mg, and other metal ions are often bonded to proteins at active sites and perform enzymatic reactions. - all prosthetic groups are added after translation. ** happens in the golgi or sER** |
|
|
Native Conformation:
|
- the shape of the protein is a balance of torque that the various interactions induce and the stress of bending the molecule.
- the native conformation will be a balance of the induced twisting and the structural stress caused by the twisting. |
|
|
How do reactive sites form on enzymes:
|
- tertiary confers a globular share on many protein, which leads to pockets and cavities in the protein which are often actives sites or regulatory sites.
|
|
|
What are the characteristics of an active sites?
|
- cavity has a specific shape which is determined by primary- tertiary interactions.
- based on enzyme's specificity - cavity fits a specific molecule(s) which is determined by the shape of the cavity and the molecule. - some of the amino acids on the active site are bound to a prosthetic group which usually has a metal ion that binds reversibly to a substrate and catalyzes a change in the substrate. - provides a micro environment for the specific reaction to take place. |
|
|
How the active site interacts with substrate:
|
1. majority of amino acids of the active site are predominantly non-polar (generally excluding water)
2. may also have some amino acids that make the site acidic. Acidic groups interact with substrate to destabilize certain bonds or a substrate 3. may have a few amino acids which are polar that are strategically placed to hold prosthetic groups which induce electrical instability in certain bonds in substrate. 4. may have a few amino acids which are polar. these amino acids are strategically placed to interact with substrates and produce physical instability in certain bond substrates [2,3,4 get the substrate to a transitional state so that it can transform into a product] |
|
|
Carbonic Anhydrase active site:
|
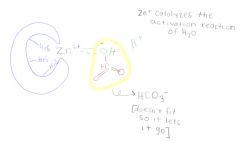
1. reaction: H2O + CO2 ---> HCO3- + H+ (bicarbonate part of carbonate buffer system of mammal blood)
2. active site must fit water and carbon dioxide together 3. The active site has 3 his residues (polar) coordinately bonded to zinc ion at bottom of a deep cleft. 4. zinc catalyses water into -OH + H+ 5. near by residues attract carbon dioxide and is held in perfect orientation to the -OH 6. electrical environment provided by Zn ion stabilizes the -OH long enough that it will be attracted to the C in the CO2, and the -OH is added to the carbon dioxide to may bicarbonate |
|
|
Lock and key model of catalysis:
|
- highly complementary shape of active site and substrate = tight fit.
- enzyme orients substrate(s) to the correct relative position, ions create the correct electrical environment and the reaction occurs making products. - products no longer fit therefore are released. - highly specific making it well able to exclude non-substrate molecules from active site. |
|
|
Induced fit model of catalysis:
|
- active site assumes shape of substrate after binding the substrate (conformational change) [ clamps down on substrate and enduces]
- conformational change puts "pressue" on certain bonds of substrate... up to the point of breaking lowering the energy of activation (E sub a) |
|
|
Cont. on induced fit model of catalysis:
|
- the kinetic energy breaks the bond and new molecule (products) are formed
- products no longer fit the active site and are expelled - the enzyme returns to its native conformation because it goes back to its lowest activation energy - these enzymes tend to be more flexible and are a bit less specific to substrate bound. may bond to several different substrates. |
|
|
Which model is more common between the lock and key vs. induced fit?
|
The induced fit model
|
|
|
What is Km?
|
a measure of how strongly a substrate is attracted and binds to its specific enzyme and then how quickly the reaction takes place and the products are expelled.
[how quickly does it happen] it is a measure of substrate turn over by a specific enzyme (turnover rate: how quickly, how well does it bind it) varies considerably among enzymes and their substrates and is affected by environmental conditions. |
|
|
How is Km measured?
|
in micro M of substrate required for enzyme to reach 1/2Vmax
|
|
|
What does lower values of Km mean?
|
the enzyme has higher binding affinity for that substrate.
[more efficient enzyme] **more like lock and key method** |
|
|
What does higher values of Km mean?
|
lower binding affinity for that substrate.
** more like induced method** |
|
|
In what ways could the enzyme mechanism be disrupted (inhibited)?
|
- removal of substrate
- non optimal: temp, solute, pH - add compounds that attach to enzyme (chemical inhibitors) |
|
|
What are some different types of chemical inhibitors?
|
1. irreversible: attach permanently to active site (or else where)= destroys enzyme
2. reversible: attach transiently to inhibit enzyme activity. |
|
|
What are the two types of reversible chemical inhibitors?
|
1. competitive inhibitors
2. allosteric effectors |
|
|
Competitive inhibitor:
|
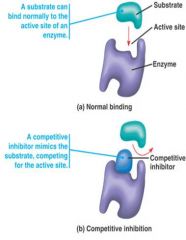
the inhibitor competes for the active site with substrate.
- inhibitor usually physically resembles the substrate but can not be changed into a product - uncommon way to control enzyme activity - this kind of inhibition can be overcome by adding more substrate |
|
|
Allosteric effectors:
|
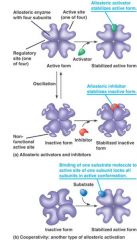
1. attaches to allosteric site and the binding changes conformation of enzyme
2. makes enzyme active or inactive 3. this kind of inhibition can not be overcome by an increase in the substrate ** binding of allosteric effectors changes the conformation of the enzyme... causing change in the enzyme's Km for the substrate. |
|
|
Lineweaver/burke plot of a competitive inhibitor:
|
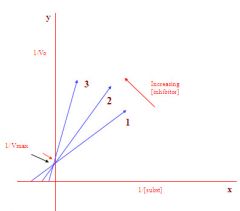
1. 1/Vmax did not change as the inhibitor increased
2. enzyme conformation did not change as a result of adding inhibitor 3. there fore must be a competitive inhibitor as this type of inhibitor does not physically change the enzyme |
|
|
Vo in a lineweaver-burke plot?
|
initial velocity of reaction
|
|
|
Km in a lineweaver-burke plot?
|
substrate concentration at 1/2 Vmax
|
|
|
Vmax in a lineweaver-burke plot?
|
max. rate at which an enzyme can function
|
|
|
Lineweaver/Burke plot of an allosteric inhibitor:
|
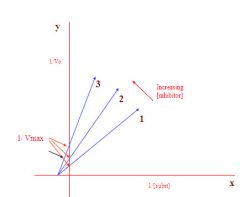
1. 1/Vmax does change as the inhibitor increases
2. the enzyme conformation did change as a result of adding inhibitor 3. therefore this must be an allosteric inhibitor as this type of inhibitor does physically change the enzyme |
|
|
Why are lineweaver/burke plots important?
|
1. allows to look at graphical data and use logic to glean info from it.
2. biochemists use this method to determine an inhibitors mode of action. |

