![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
75 Cards in this Set
- Front
- Back
|
chromatin:
|
- DNA-protein complex
- packs a cells DNA into a compact form that fits inside the nucleus - important in helping regulate gene expression. |
|
|
Histones:
|
are proteins responsible for the first level of DNA packing in chromatin.
- have a high proportion of positively charged amino acids and they bind tightly to the negatively charged DNA. |
|
|
Nucleosome:
|
the "bead"
- basic unit of DNA packing - consist of DNA wound around a protein core composed of two molecules each of 4 types of histone: H2A, H2B, H3, H4 |
|
|
Linker DNA:
|
the "string" between the bead
|
|
|
How can DNA be transcribed when it is wrapped around histones in a nucleosome?
|
the change in the shapes and positions of nucleosomes can allow RNA-synthesizing polymerases to move along the DNA
|
|
|
Heterochromatin:
|
- irregular clumps
- DNA is largely inaccessible to transcription enzymes - generally is not transcribed |
|
|
euchromatin:
|
- "true chromatin"
- looser packing of euchromatin makes its DNA accessible to enzymes & available for transcription. |
|
|
Describe the structure of a nucleosome, the basic unit of DNA packing in eukaryotic cells:
|
made up of 8 histone proteins, two each of 4 different kinds, around which DNA is wound. Linker DNA runs from one nucleosome to the next one.
|
|
|
What chemical properties of histones & DNA enable these molecules to bind tightly together?
|
histones contain many basic amino acids (+ charged), which can form weak bonds with the neg. charged phosphate groups on the sugar-phosphate backbone of the DNA molecule
|
|
|
In general, how does dense packing of DNA in chromosomes prevent gene expression?
|
RNA polymerase and other proteins required for transcription to occur do not have access to the DNA in tightly packed regions of a chromosome.
|
|
|
Levels of Chromatin Packing:
|
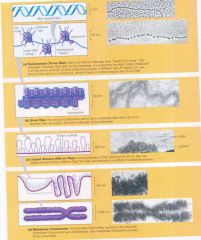
This sereis of diagrams and transmission electron micrographs depict a current model for the progressive stages of DNA coiling and folding.
|
|
|
Cell Differentiation:
|
- cells undergo a development of a multicellular organism
- process of specialization - results in several/many differentiated cell types. |
|
|
Differential gene expression:
|
- the expression of different genes by cells with the same genome
- explains the differences between cell types. |
|
|
Stages in gene expression that can be regulated in eukaryotic cells:
|

In this diagram, the colored boxes indicate the processes most often regulated. The nuclear envelope separating transcription from translation in eukaryotic cells offers an opportunity for post-translation in eukaryotic cells offers an opportunity for post-transcriptional control in the form of RNA processing that is absent in prokaryotes. In addition, eukaryotes have a grater variety of control mechanisms operating before transcription and after translation. The expression of any given gene, however, does not necessarily involve every stage shown
|
|
|
Chemical modificaitons to histone play a direct role in the regulation of what?
|
gene transcription.
|
|
|
Histone acetylation:
|
- acetyl groups (-COCH3) are attached to positively charged lysines in histone tails.
- + charges are neutralized therefore no longer bind to neighboring nucleosomes (causing chromatin to have a looser structure) results: transcription proteins have easier access to genes in an acetylated region. |
|
|
A simple model of histone tails and the effect of histone acetylation
|
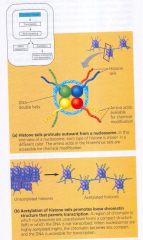
In addition to acetylation, histones can undergo several other types of modifications that also help determine the chromatin configuration in a region.
|
|
|
Methylation:
|
- the addition of a methyl group (-CH3)
- lead to condensation of the chromatin. |
|
|
Histone code hypothesis:
|
specific combinations of modifications, rather than the overall level of histone acetylation, help determine the chromatin configuration which in turn influences transcription.
|
|
|
Epigenetic Inheritance:
|
inheritance of traits transmitted by mechanisms not directly involving the nucleotide sequence.
|
|
|
Function of chromatin-modifying enzymes:
|
provide initial control of gene expression by making a region of DNA either more or less able to bind the transcription machinery.
|
|
|
A eukaryotic gene & the DNA elements that control it:
|
- cluster of proteins (a transcription initiation complex) assembles on teh promoter sequence at the "upstream" end of the gene.
- RNA polymerase II, proceeds to transcribe the gene, synthesizing a primary RNA transcript (pre-mRNA) - RNA processing includes enzymatic addition of a 5'cap & a poly-A tail as well as splicing out of introns to yeild a mature mRNA. |
|
|
Control elements:
|
- segments of noncoding DNA that help regulate transcription by binding certain proteins.
- critical to the precise regulation of gene expression. |
|
|
A Eukaryotic gene and its transcript.
|
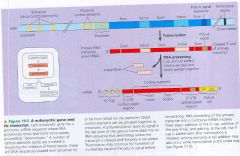
|
|
|
Transcription factors:
|
proteins which assist RNA polymerase to initiate transcription.
|
|
|
The interaction between what are particularly important in controlling gene expession?
|
interactions between enhancers and specific transcription factors call activators or repressors.
|
|
|
activator:
|
a protein that binds to an enhancer and stimulates transcription of a gene.
|
|
|
repressors:
|
specific transcription factors which inhibit expression of a particular gene.
|
|
|
A model for the action of enhancers and transcription activators:
|
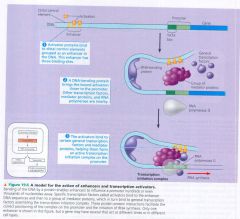
|
|
|
Cell Type-specific transcription
|

|
|
|
How does the eukaryotic cell deal with genes of related function that need to be turned on/off at the same time?
|
- each eukaryotic gene in these clusters has its own promoter & is individually transcribed (co-expressed genes clustered near one another on the same chromosome)
------> The coordinate regulation of clustered genes is thought to involved changes in the chromatin structure that makes the entire group of genes either available/unavailable for transcription. - coordinate control of dispersed genes often occur in response to external chemical signals. |
|
|
How is the expression of a protein-coding gene ultimately measured?
|
by the amount of functional protein a cell makes & much happens between the synthesis of the RNA transcript & the activity of the protein in the cell.
- increasing # of regulatory mechanisms that opperate @ various stages after transcription allow a cell to fine-tune gene expression rapidly in response to environmental changes w/o altering its transcription patterns. |
|
|
Alternative RNA splicing:
|
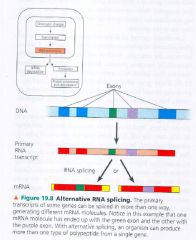
- an example of regulation at the RNA-processing level
- different mRNA molecules are produced fromt eh same primary transcript, depending on which RNA segments are treated as exon & which are introns. |
|
|
What is an important factor in determining the pattern of protein synthesis in a cell?
|
the life span of mRNA molecules in the cytoplasm.
- mRNA typically survive for hrs, days, or even wks. |
|
|
microRNAs (miRNAs):
|
- mechanism that blocks expression of specific mRNA
- small single-stranded RNA molecules - can bind to complementary sequences in mRNA molecules. - are formed from long RNA precursors that fold back on themselves, forming a long, double-stranded hairpin structure held together by H-bonds. - an enzyme, fitting (Dicer) cuts the fragments one strand degrades, the other is the miRNA. - miRNA complex either degrades the target mRNA or blocks its translation. |
|
|
Regulation of gene expression by microRNAs (miRNAs):
|
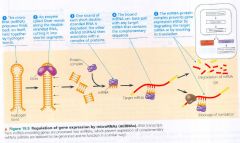
|
|
|
RNA interference (RNAi):
|
- due to small interfering RNA (siRNAs)
- the inhibition of gene expression -----> the injecting double-stranded RNA molecules into cell somehow turned off a gene w/ the same sequence. |
|
|
small interfering RNAs (siRNA):
|
similar size & funciton as miRNAs
|
|
|
What are some processed polypeptides go through to yield functional protein molecules?
|
- cleavage of initial polypeptides forms active hormone
- undergo chemical modification that makes them functional - activated/ inactiviated by the reversible addition of phosphate groups - transported to targeted destionation. |
|
|
proteasomes:
|
- giant protein complexes
- recognize the ubiquitin-tagged protein molecules and degrade them. |
|
|
Degradation of a protein by a proteasome:
|
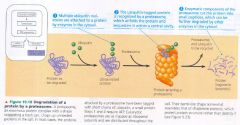
|
|
|
In general, what is the effect of histone acetylation and DNA methylation in gene expression?
|
- histone acetylation is generally associated with gene expression.
- DNA methylation is generally associated with lack of expression. |
|
|
compare the roles of general & specific transcription factors in regulating gene expression.
|
- general transcription factors function in assembling the transcription initiation complex at the promoter for all genes.
- specific transcription factors bind to control elements associated with a particular gene and once bound either increase (activators) or decreases (repressors) transcription of that gene. |
|
|
if you compared the nucleotide sequences of the distal control elements in the enhancers of three corrdinately regulated genes, what would you expect to find? why?
|
the 3 genes should have some similar/identical sequences in the control elements of their enhancers. because this similarity, the same specific transcription factors could bind to the enhancers of all 3 genes & stimulate their expression coordinately.
|
|
|
once mRNA encoding a particular protein reaches the cytoplasm, what are 4 mechanisms that can regulate the amount of the active protein in the cell?
|
1. degradation of the mRNA
2. regulation of translation 3. activation of the protein 4. protein degradation. |
|
|
What accounts for 24% of the human genome?
|
gene-related regulatory sequence and introns
|
|
|
Repetitive DNA:
|
sequences that are present in multiple copies in the genome
- about 3/4 of this DNA (44% of the entire human genome) is made up of transposable elements & sequences related to them. |
|
|
Transposons:
|
- move w/in a genome by means of a DNA intermediate
- can move by a "cut & paste" mechanism, which removes the element from the original site or by a "copy & paste" mechanism, which leaves a copy behind. |
|
|
retrotransposons:
|
- move by means of an RNA imtermediate.
- always leave a copy behind at the original site. |
|
|
Movement of eukaryotic transposable elements:
|
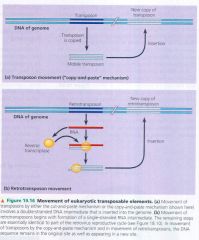
|
|
|
Satellite DNA:
|
- simple sequence DNA
- has different density that the rest of the cell's DNa - repetitive DNA - called satellite DNA b/c it appeared as a "satellite" band in the centrifuge tube - located at chromosomal telomeres & centromeres -------> suggest DNA plays a structural role for chromosomes. |
|
|
DNA at centomeres:
|
essential fro the separation of chromatids in cell division.
|
|
|
DNA at telomeres:
|
at the tips of chromosomes, prevents genes from being lost as the DNA shortens with each round of replication.
- also binds proteins that protect the ends of a chromosome from degradation and from joining to other chromosomes. |
|
|
The sequences coding for proteins & structural RNAs compose about how much % in the human genome?
|
about 1.5%
|
|
|
multigene families:
|
collection of identical / very similar genes.
|
|
|
example of gene families:
|

|
|
|
Pseudogenes:
|
nonfunctional nucleotide sequences quite similar to the functional genes.
|
|
|
Discuss the characteristics that make mammalian genomes larger than prokaryotic genomes.
|
the # of genes is 5-15 times higher in mammals, & the amount of noncoding DNA is about 10,000 times greater.
the presence of introns in mammalian genes make them about 27 times longer, on average, than prokayotic genes. |
|
|
how do introns, transposable elements & simple sequence DNA differ in their distribution in the genome?
|
Introns:
are interspersed w/in the coding sequences of genes. Transposable elements: many copies of transposable elements are scattered throughout the genome. simple sequence DNA: is concentrated at the centromers and telomeres. |
|
|
discuss the differences int eh organization of the rRNA gene family and the globin gene families. How do these gene families benefit the organism?
|
rRNA gene family:
- idential transcription units encoding three different RNA product are present in long, randomly repated arrays. - the larger # of copies of the rRNA genes enable organisms to produce the rRNA for enough ribosomes to carry out active protein synthesis. globin gene families: - consists of relatively small # of nonidentical genes clustered near each other. - the difference in the globin proteins encoded by these genes result in teh production of hemoglobin molecules adapted to particular developmental stages of the organism. |
|
|
eu- =
|
true
|
|
|
hetero- =
|
different
|
|
|
nucleo- =
|
the nucleus
|
|
|
-soma=
|
body
|
|
|
proto- =
|
first, original
|
|
|
onco- =
|
tumor
|
|
|
pseudo- =
|
false
|
|
|
retro- =
|
backward
|
|
|
The control of gene expression is more complex in eukaryotic cells because
a) DNA is associated with protein b) gene expression differentiates specialized cells c) the chromosomes are linear and mroe numerous d) operons are controlled by more than one promoter region e) inhibitory or activating molecules may help regulate transcription. |
b) gene expression differentiates specialized cells.
|
|
|
histones are
a) small, positively charged protein that bind tightly to DNA b) small bodies in the nucleus involved in rRNA synthesis. c) basic units of DNA packing consisting of DNA wound around a protein core d) DNA bending proteins that facilitate formation of transcription initiation complexes. e) proteins responsible for producing repeating sequences at telomeres. |
a) small, positively charged proteins that bind tightly to DNA
|
|
|
Heterochromatin:
a) has a higher degree of packing than does euchromatin b) is visible with a light microscope during interphase. c) is not actively involved in transcription d) makes up metaphase chromosomes e) is all of the above |
e) is all of the above
|
|
|
Which of the following is not true of enhaners?
a) they may be located thousands of nucleotides upstream from the genes they affect. b) when bound with activators, they interact with the promoter region and other transcription factors to increase the activity of a gene. c) They may complex with steroid-activated receptor proteins and thus selectively activate specific genes. d) they may coordinate the transcription of enzymes involved in the same metabolic pathway. e) they are located within the promoter, and when complexed with a steroid or other small molecule, they release an inhibitory protein and thus make DNA more accessible to RNA polymerase. |
e) they are located within the promoter, and when complexed with a steroid or other small molecule, they release an inhibitory protein and thus make DNA more accessible to RNA polymerase.
|
|
|
Which of the following is not an example of the control of gene expression that occurs after transcription?
a) mRNA stored in the cytoplasm needing activation of translation initiation factors b) the length of time mRNA lasts before it is degraded c) rRNA genes amplified in tandem arrays d) alternative RNA splicing beofore mRNA exits from the nucleus e) splicing or modification of a polypeptide |
c)rRNA genes amplified in tandem arrays
|
|
|
Pseudogens are?
a) tandem array of rRNA genes that enable actively synthesizing cells to create enough ribosomes b) genes that can become oncogenes when mutated by carcinogens c) genes of multigene families that are expressed at different times during development d) sequences of DNA that are similar to real genes but lack regulatory sequences necessary for gene expression e) both c and d |
d) sequenes of DNA that are similar to real gens but lack regulatory sequences necessary for gene expression.
|
|
|
What are proteasomes?
a) complexes of proteins that excise introns b) single-stranded RNA moleculs complexed with proteins that block translation of or degrade mRNA c) small, positively charged proteins that form the core of nucleosomes d) enormous protein complexes that degrade unneeded proteins in the cell e) complexes of transcription factors whose protein-protein interactions are required for enhancing gene transcription. |
d) enormous protein complexes that degrade unneeded proteins in the cell.
|

