![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
91 Cards in this Set
- Front
- Back
|
Amphiphilic / Amphipathic
|
Have one end that will dissolve in water and one end that wil disoolve in a nonpolar environment
|
|
|
Hydrogen Bonds
|
Hydrogen atom covalently bonded to one electronegative atom interacts with another electronegative atom
|
|
|
In SDS-PAGE, separation takes place on the bases of:
|
The sieving action of the gel, since all particles have approx the same charge/mass ratio, but dif. masses
|
|
|
A single water molecule can participate in up to __ H-bonds
|
4
|
|
|
The dominant interaction that drives a water-soluble protein to fold is:
|
The hydrophobic interaction
|
|
|
Which substance would be a suitable buffer at pH 5.0?
|
one with a pKa of ~ 5.0
|
|
|
Two amino acids frequently found in reverse turns are:
|
glycine and proline
|
|
|
In isoelectric focusing gel electrophoresis:
|
there is a pH gradient that parallels the electric fild gradient
|
|
|
True statements about water and/or H-bonds
|
1. H-atoms of water each bear a partial pos charge
2. H-bonds can form b/w diff parts of polypeptide chain 3. H-bonds are relatively weak compared to covalent bonds. 4. H-bonds in water contribute to its cohesiveness |
|
|
Procedure necessary to test whether amino acid sequence of ibonuclease contains al the information needed for it to fold into its native, fucnal 3-d structure
|
1. FIRST: add 8M urea; RESULT: denatures, partially unfolds protein
2. FIRST: add reducing agent; RESULT: breaks S-S, RNase now completely unfolds 3. FIRST: dialyze away urea; RESULT: removal of denaturant, partial re-folding 4. FIRST: gendle air oxidation; RESULT: disulfides reform, final correct folding, yielding native form |
|
|
Find the pH of 1L solution containing 0.52M sodium acetate and 0.48M acetic acid. The pKa of acetic acid is 4.76
|
pH = pKa + log[A-]/[HA]
= 4.76 + log 0.52/0.48 = 4.76 + 0.035 pH = 4.79 |
|
|
Find the pH of a solution after addition of 0.04 moles of HCl. Assume that HCl addition causes no change in volume.
|
HA <===> H+ + A-
[A-] = 0.52 - 0.04 = 0.48 [HA] = 0.48 + 0.04 = 0.52 pH = pKa + log0.48/0.52 = 4.76 - 0.035 pH = 4.73 |
|
|
Non-Covalent Interactions
|
1. H-bonds: b/w polar groups
2. Electrostatic (ionic): b/w charged groups 3. Hydrophobic: among non- polar groups in aq. solut'n 4. Van Der Waals |
|
|
Delta G0
|
Standar Free Energy: Constant based on standard conditions --> additive for seuential reactions
|
|
|
Delta G
|
Actual Free energy change for eaction and function of concentration of products and reactant; fcn of temperature
|
|
|
Ionizable Protein Groups
|
1. terminal a-carboxyl group
2/3. aspartic & glutamic acid 4. Histidine 5. terminal a-amino group 6. Cysteine 7. Tyrosine 8. Tyrosine 9. Lysine 10. Arginine |
|
|
pKa of terminal a-carboxyl group
|
3.1
|
|
|
pKa of Aspartic/Glutamic acid
|
4.1
|
|
|
pKa of Histidine
|
6.0
|
|
|
pKa of terminal a-amino group
|
8.0
|
|
|
pKa of Cysteine
|
8.3
|
|
|
pKa of Lysine
|
10.8
|
|
|
pKa of Tyrosine
|
10.9
|
|
|
pKa of Arginine
|
12.5
|
|
|
Gobular proteins
|
Gobular proteins are involved in the chemical functions of living things. The ones known as enzymes are organic catalyses that speed up the chemical reaction rates in cells. Other important gobular proteins are hemoglobin, antibodies, and some hormones.
|
|
|
Primary structure
|
There are four levels of structure in proteins. The simplest is level is called primary structure. It is determined by the kind, number and sequence of amino acids. The amino acids or peptides are joined to one another by strong covalent bonds. These bonds form between the nitrogen atom of the amino group and the oxygen atom of the carboxyl group. They are called peptide bonds.
|
|
|
Secondary structure
|
The second level of structure involves the coiling of primary chains into a helix or the formation of what are called pleated (beta) sheets Collagen is an example of a pleated sheet. Bonding between sulfhydryl groups of the amino acid cystine forms a strong covalent bond called a disulfide bond. In addition, ionic and weak hydrogen bonds help to stabilize the secondary structure.
|
|
|
Tertiary structure
|
The third level of protein structure has increased folding between alpha helix and non alpha helix regions. The structure is stabilized by disulfide, ionic, and hydrogen bonding. Gamma gobulin is an example of a gobular protein exhibiting tertiary structure. Keratin is a fibrous protein found in animal hair, claws and fingernails.
|
|
|
Quaternary structure
|
The final level of protein structure is composed of sub units of polypeptide chains with or without non amino acids. Like the two levels before them they are held together by disulfide, ionic and hydrogen bonding. Hemoglobin exhibits this structure. It is also an example of a conjugated protein because of the presence of a non amino acid group called the "heme group".
|
|
|
Denaturating of a Protein
|
The term denaturation is used to describe changes in a protein's shape. Such changes maybe temporary or permanent. In either case the breaking of the stabilizing bonds of a protein leads to its inactivation.
Any of the following conditions can bring about the denaturing of a protein: * heat * pH * pressure * chemicals * heavy metals |
|
|
Hydrophobic "Effect"
|
Other molecules exclude water molecules b/c of organization necessary to deal w/ H-bonds.
|
|
|
Hydrophobic "Effect" in Relation to DNA
|
Hydrophobic effect stabilizes stacking of base pairs in a linear piece of DNA;
Backbone (sugar) of DNA ar neg. charged, Electrostatic forces keep them apart and neutralized |
|
|
Zwitterionic Form
|
IN NEUTRAL pH!
Amino acids at neutral pH exist mostly as dipolar ions -> amino is protonated (--NH3+) -> carboxyl is de (--COO-) |
|
|
Amino Acids in Acidic Solution
|
-> amino is prot'd (--NH3+)
-> carboxyl is prot'd (--COOH) |
|
|
Amino Acids in Basic Solution
|
-> amino is deprotonated (--NH2)
-> carboxy is also de (--COO-) |
|
|
Directionality of A.A. sequence
|
AMINO -> -> CARBOXYL
(N-term) (C-term) |
|
|
Metabolite
|
Low molecular weight molecule such as glucose and glycerol
|
|
|
Epigenetic Factors
|
Associated w/ the genome by not represented in sequence of DNA
|
|
|
Dalton
|
A unit of mass nearly = to that of a hydrogen atom
|
|
|
B-Pleated Sheet Stabilization & Bonds
|
* Stabilized by H-bonds b/w polypeptide strands
* Distance b/w AA = 3.5A * Side chains of AAs point in opposite directions |
|
|
Antiparallel B-Sheets
|
Adjacent B-sheets run in opposite direction
-> H-Bonds form b/w NH & CO groups; connect each AA to a single AA on an adjacent strand |
|
|
Parellel B-Sheets
|
Adjacent B-sheets run in the same direction
-> H-bonds connect each AA on one strand to TWO different AA on another strand * slightly less stable than antiparallel b/c of H-bond angle strain |
|
|
pKa of carboxyl group
|
3.1
|
|
|
pKa of aspartic acid / glutamic acid
|
4.1
|
|
|
pKa of histidine
|
6.0
|
|
|
pKa of amino group
|
8.0
|
|
|
pKa of cysteine
|
8.3
|
|
|
pKa of lysine
|
10.8
|
|
|
pKa of tyrosine
|
10.9
|
|
|
pKa of arginine
|
12.5
|
|
|
Phi angle
|
Torsion angle between a-carbon and nitrogen
|
|
|
Psi angle
|
Torsion angle between a-carbon and carboxyl carbon
|
|
|
(D)extroratotory
(L)evoratatory |
D-Right // L-Left
* Refers to plane polarized light; uniform w/ respect to Z dimension; L vs D is determined by passing p.p. light through solution of molecule |
|
|
a-Keratin
|
Primary component of wool and hair.
* Consists of 2 right-handed a-helices intertwined to form a left handed helix: a-coiled coil * Two helices are cross-linked by van der waals (if repeated residues are hydrophobic) & ionic (if repeated residues are hydrophilic) & disulfide (from cysteine) * # of cross-links determine flexability |
|
|
Collagen Helix
|
Rod-shaped; 3-helical polypeptide chain; GLY every 3 residues
* no H-bonds, instead stabilized by steric interactions/repulsion |
|
|
Denatured Protein
|
Protein converted into a randomly coiled peptide w/o its normal activity
|
|
|
Sequence Specifies Conformation
|
The information needed to specify catalytically active structure of ribonuclease is contained in its amino acid sequence -> this structure is generally thermodynamically favored.
|
|
|
Chaperone Proteins
|
Proteins inside cells that prevent wrong folds under normal conditions
-> get energy from ATP |
|
|
When will a protein conform to an a-helix?
|
"Default" Conformation
-> esp sequences including * ALA * GLU * LEU |
|
|
When will a protein conform to a b-sheet?
|
* Residues w/ VAL, THR, ILE, because branching disrupts a-helix
* Also when side chains contain H-donors * acceptors that interact w/ main chain |
|
|
When will there be a reverse turn?
|
Generally, when there's a proline -> ring structure is restrictive to a-helix or b-sheet.
|
|
|
Cooperative Transition
|
Proteins tend to be either all folded or all unfolded
-> protein placed in condition where some part is thermodynamically unstable: part of folded structure is disrupted -> interact'ns b/w it and remainder of protein are lost -> destabilizes entire protein |
|
|
Leventhal's Paradox
|
Difference b/w calculated and actual folding time of a protein
-> Reveals that proteins do not fold by trying every conformation |
|
|
Cumulative Selection of Protein Folding
|
* Partly correct intermediates are retained
-> correctness is not based on residues per se, but on total free energy -> proteins are only marginally stable |
|
|
Nucleation Condensation Model (Perscht) of Protein Folding
|
* A difuse folding nucleus is formed & consolidated through the transition state
-> local regions taht have significant structural preference, though not nec. stable on their own, will tend to adopt their favored structure ->interact: increase stability * Viewed as a funnel: begins w/ highest free energy & many possible conformations; as free energy decreases, so do possible accessible conformations |
|
|
Insulin
|
Triggers phosphorylation of -OH of TYR
|
|
|
Matthews ('95)
|
Relationship b/w protein stability & protein function
* showed by residue substitution that increased stability reduces activity. -> proteins fold to minimize free energy -> organize themselves to recognize ligand |
|
|
How is Stability of Protein Achieved?
|
Hydrophobic inside
Hydrophilic outside |
|
|
Priority of Functional Groups
|
SH > OH > NH2 > COOH > CHO > CH2OH > C6H5 > CH3 > H
|
|
|
Anfinsen (1952)
|
First demonstrated that primary structure determines 3-D shape
* RNaseA (really hard to denature) & urea to denature & DTT to reduce disulfide bonds -> showed RNaseA could complex w/RI -> then used dialysis to renature -> regained activity |
|
|
Proteome
|
represents the fcnal expression of information, such as type, fcn, & interactions w/ protein that yield as a fcnal unit
|
|
|
Salting Out (precipitation)
|
Can be used to fractionate proteins since most are less soluble at high [salt]. This concentration (at which protein is no longer soluble) is diff for diff proteins
-> use [diff] depending on what you want to precipitate |
|
|
Dialysis
|
Semipermeable membrane -> proteins too big to leave bag, while small molecules may leave
* useful to get rid of salt, although it won't sep diff proteins |
|
|
Gel Filtration Chromatography
|
SIZE: LARGE=FAST, SMALL=SLOW
* gel (such as agarose) contains many beads, large molecules flow around them, small molecules get stuck in them |
|
|
Ion Exchange Chromatography
|
CHARGE: (+) = SLOW, (-)=FAST
* gel contains neg charged beads (which contain carboxylate group); pos proteins will bind and take longer to run. (can also contain beads w/ (+) charge) |
|
|
Affinity Chromatography
|
FUNCTIONAL GROUPS
* separat'n based on a highly specific biologic interaction such -> antigen and antibody -> enzyme and substrate -> receptor and ligand. |
|
|
HIS-tags
|
Proteins containing HIS tags are passed through a column of beads containing covalently attached, immobilized nickel (II) or other metal ions
-> HIS-tag binds tightly to immobilized metal ions, binding desired protein while other proteins flow though the column * protein eluted by addition of imidazole or another molecule that displaces his |
|
|
HPLC
|
* similar to other column techniques but column materials are much finer
-> more interaction sites = greater resolving power -> pressure added to obtain adeq. flow rate |
|
|
Gel Electrophoresis
|
NET CHARGE AND SIZE
* run on polyacrylamide gel velocity=Ez/f f-depends on mas and shape and viscosity of medium) z=charge E=electric field strength |
|
|
Direction to run Gel Electrophoresis
|
RUN TO RED
(-) --> (+) |
|
|
To separate based on mass only
|
1. SDS (anionic detergent): denatures by disrupting noncovalent interactions
2. mercaptoEtOH: reduces disulfide bonds 3. Anion of SDS binds to main chain * Complex now has large net (-) charge -> runs based on mass * separate w/electrophoresis * visualize w/coomasie blue * if radioactively labeled, arg (x-ray film over gel) |
|
|
Acrylamide
|
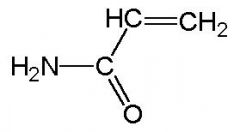
Acrylamide
|
|
|
Why do we use a detergent?
|
Provides uniform charge to the protein (makes all neg.) by binding to peptide backbone stoichiometrically -> all proteins run towards (+), not back off the gel
|
|
|
Detergent used in SDS-PAGE
|
sodium dodecyl sulfate
O " Na+ -O-S-O-(CH2)-CH3 " O |
|
|
Names Helices
|
#residues/turn followed by # atoms in ring formed by H-bond
ex:a-helix=3.6 13 3 10 =tighter, narrower, longer |
|
|
Turns
|
* Pretty rigid
* 4AA, often contain PRO & GLY * small * simply there to change direction |
|
|
Loops
|
* bigger
* often flexible, sometimes not visible on x-ray |
|
|
Induced Fit
|
often unstructured loop will acquire structure when interactions w/ another protein
|
|
|
Protein Domain
|
Series of discrete secondary structure in a singe peptide chain (~string of pearls)
-> often domains have specific/unique fcn or can fold independently |
|
|
Isoelectric Focusing
|
CONTENT OF ACIDIC & BASIC RESIDUES
* establish pH gradient & add voltage * proteins stop moving at their isoelectric point (pH at which net charge is zero) |
|
|
Overexpression System
|
Grow protein in non-native (foreign) organism
-> hRI is produced in E.coli * Plasmid uses genetic elements from nature to control expression |

