![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
16 Cards in this Set
- Front
- Back
|
Transcription
|
The synthesis of a ssRNA copy of a segment of DNA
|
|
|
Basic Process of Transcription
|
1) DNA unwinds in a short region
2) RNA polymerase catalyzes the synthesis of an RNA in the 5' to 3' direction by reading 3' to 5' Ribonucleoside triphosphates are the RNA precursor molecules used for RNA synthesis 3) RNA polymerase have ability to synthesize WITHOUT primer |
|
|
Antisense and Sense Strand
|
Is the template strand of DNA that will be used for RNA synthesis. The sense strand will be the other DNA strand, and will have thee same sequence as the RNA synthesized.
|
|
|
Comparision of DNA and RNA Syntehesis
|
RNA:
RNA polymerase NTPs precursor (ribonucleoside triphosphates) no primer needed uracil pairs with adenine DNA: DNA polymerase dNTP precursor (deoxyrobonucleoside triphosphate) primer required for initiation thymine pairs with adenine |
|
|
mRNA
|
encodes the amino acid sequence of a polypeptide. They are the transcripts of a protein coding genes.
|
|
|
tRNA (transfer RNA)
|
Brings the amino acids to the ribosomes during the translation process
|
|
|
rRNA (ribosomal RNA)
|
Combines with ribosomal proteins to form the ribosome. mRNA is translated into protein at the ribosome
|
|
|
snRNA (small nuclear RNA)
|
Combines with certain proteins and is and is involved in RNA processing (mRNA splicing) and gene regulation
|
|
|
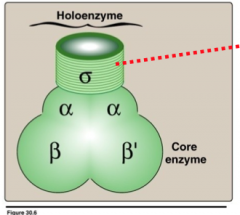
RNA Polymerase in bacteria
|
There is only only a single RNA polymerase. Associates with protein called sigma factor. Together are called holozyme, the sigma factor recognizes place where transcription starts called a promoter.
Core enzyme has helicase and polymerase activity |
|
|
RNA Polymerases in Eukaryotes
|
RNA Pol1: involved in synthesizing rRNA 5.8S, 18S and 28S
RNA Pol2 2: involved in synthesizing mRNA, some snRNA and miRNA RNA Pol 3: synthesizes tRNA and some snRNA |
|
|
Promoter and Terminator
|
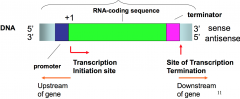
Located upstream of RNA coding sequence. Where rNA polymerase will bind to before starting transcription.
Termination sequence that tells RNA polymerase will to stop |
|
|
Promoter
|
Prokaryote promoter are found at -35 and -10 bp from transcription site:
-10 sequence: called Pribnow box This RNA polymerase and sigma factor bind between these two sites, allow itself to position so it starts at +1. |
|
|
Steps of Transcription
|
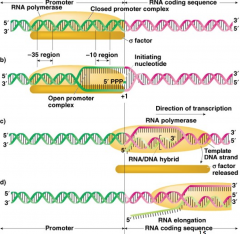
1) RNA polymerase and sigma factor will bind between -35 and Pribnow sequence. RNA Pol has its own helicase and will begin to unwind DNA about 16 bases in the -10 region. Starts synthesis, no primer needs
2) Elongation: Syntehsizes RNA in the 5' to 3'. Will synthesize 8-9 bases initially and sigma factor disassociates from core enzyme. Sigma only needed for initial binding and synthesis. 3) Chain termination and release of RNA. Reaches termination sequence and stops and separates |
|
|
Rifampin (rifomyocin)
|
-Antibiotic used in tuberculosis
-inhibits transcription -blocks formation of the first phosphodiester bind by binding to prokaryote RNA polymerase (of eukaryotes not prokaryotes). Binds to beta subunits because of shape and fit (lock and key) -specific base -Mutations in genes encoding for RNA polymerase can lead to drug resistance |
|
|
Eukaryotic Promoters
|
GC Box: -70 to -200
TATA Box: -20 to -35 CAAT box: -80 Promoters are much more complex in eukaryotes. The positioning of these promoters are variable. |
|
|
Transcription Factors
|
TATA Box binding protein, TFIID, TFIIB, TFIIH, TFIIE
|

