![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
90 Cards in this Set
- Front
- Back
|
Describe Structure of DNA
|
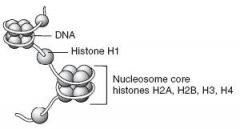
DNA exists in the condensed, chromatin form in order to fit into the nucleus. Negatively charged DNA loops twice around positively charged histone octamer(2 sets of H2A, H2B, H3, and H4) to form a nucleosome "bead".
|
|
|
What amino acids are histone octamer subunits generally comprised of?
|
Octamer subunits consist primarily of lysine and arginine amino acids, which are positively charged.
|
|
|
Which histone lies outside of the nucleosome?
|
H1 ties nucleosome beads together in a string.
Notably, H1 is the only histone that is not part of the nucleosome core. |
|
|
Heterochromatin
|
Condensed, transcriptionally inactive, sterically inaccessible.
HeteroChromatin = Highly Condensed |
|
|
Euchromatin
|
Less condensed, transcriptionally active, sterically accessible.
Eu=true, "truly transcribed." |
|
|
Nucleotides -
Purines Notable fact(s) about the purines... |
Adenine, Guanine - 2 rings
Guanine has a ketone PURe As Gold: Purines |
|
|
Nucleotides -
Pyrimidines notable fact(s) about the pyrimidines... |
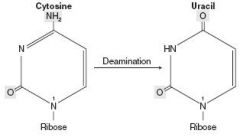
Cytosine, Thymine, Uracil - 1 ring
THYmine has a meTHYl Deamination of cytosine makes uracil. CUT the Py: pyrimidines |
|
|
Which nucleotide bond is stronger?
|
G-C. It has 3 hydrogen bonds while A-T only has 2 hydrogen bonds.
|
|
|
Requirements for purine synthesis
|
Amino Acids: Glycine, Aspartate, Glutamine
also, Tetrahydrofolate |
|
|
Nucleotide vs Nucleoside
How are nucleotides linked? |
Nucleoside = base + ribose
Nucleotide = base + ribose + phosphate Nucleotides are linked by 3'-5' phosphodiester bonds. |
|
|
FLASHCARD TO REMIND YOU TO REVIEW PURINE AND PYRIMIDINE SYNTHESIS PATHWAYS
|
REMIND YOU TO REVIEW PURINE AND PYRIMIDINE SYNTHESIS PATHWAY
|
|
|
Hydroxyurea
|
inhibits ribonucleotide reductase
|
|
|
6-mercaptopurine(6-MP)
|
blocks de novo purine synthesis
|
|
|
5-fluorouracil(5-FU)
|
inhibits thymidylate synthase. Decreases dTMP.
|
|
|
Methotrexate(MTX)
|
inhibits dihydrofolate reductase
Decreases dTMP |
|
|
Trimethoprim
|
inhibits bacterial dihydrofolate reductase
decreases dTMP |
|
|
Orotic Aciduria
Definition? Cause? Findings? Treatment? |
Inability to convert orotic acid to UMP(de novo pyrimidine synthesis pathway) due to defect in either orotic acid phosphoribosyltransferase or orotidine 5'-phosphate decarboxylase. Autosomal recessive.
Findings: increased orotic acid in urine, megaloblastic anemia(does not improve with administration of Vitamin B12 or folic acid), failure to thrive. No hyperammonemia(compared to OTC deficiency in which case you have increased orotic acid with hyperammonemia) Treatment: Oral uridine administration |
|
|
Adenosine Deaminase(ADA) Deficiency
|
Excess ATP and dATP imbalances nucleotide pool via feedback inhibition of ribonucleotide reductase which leads to prevention of DNA synthesis and thus decreases lymphocyte count. One of the major causes of SCID
|
|
|
Severe Combined Immunodeficiency Disease
|
Happens to kids(like bubble boy). 1st disease to be treated by experimental human gene therapy.
|
|
|
Lesch-Nyhan Syndrome
Definition? Cause? Findings? |
Defective purine salvage owing to absence of HGPRT, which converts hypoxanthine to IMP and guanine to GMP. Results in excess uric acid production.
Cause: X-linked recessive Findings: retardation, self-mutilation, aggression, hyperuricemia, gout, choreoathetosis He's Got Purine Recovery Trouble |
|
|
Transition
|
Substituting purine for purine or pyrimidine for pyrimidine
TransItion = Identical types |
|
|
Transversion
|
Substituting purine for pyrimidine or vice versa
TransVersion = ConVersion between types |
|
|
Features of genetic code
|
Unambiguous - Each codon specifies only one amino acid
Degenerate/Redundant - More than 1 codon may code for the same amino acid. Ex: Methionine encoded by only 1 codon(AUG). Commaless, non-overlapping - read from a fixed starting point as a continuous sequence of bases.(some viruses are an exception) Universal - Genetic code is conserved throughout evolution.(Exceptions include mitochondria, archaebacteria, Mycoplasma, and some yeasts. |
|
|
Mutations in DNA -
Silent |
Same amino acid, often base change in third position of codon(tRNA wobble).
|
|
|
Mutations in DNA -
Missense |
Changed amino acid(conservative - new amino acid is similar in chemical structure).
|
|
|
Mutations in DNA -
Nonsense |
Change resulting in early stop codon
STOP THE NONSENSE |
|
|
Mutations in DNA -
Frame shift |
Change resulting in misreading of all nucleotides downstream, usually resulting in a truncated, nonfunctional protein.
|
|
|
Grade the severity of types of mutations in DNA
|
frame shift > nonsense > missense > silent
|
|
|
DNA replication -
Rough comparison of prokaryotic vs eukaryotic DNA replication |
Eukaryotic DNA replication is more complex than the prokaryotic process but utilizes many of the same enzymes. In both cases, DNA replication is semi-conservative and involves both continuous and discontinuous(Okazaki Fragment) synthesis. For eukaryotes, replication begins at a consensus sequence of base pairs.
|
|
|
DNA Replication -
Origin of Replication |
Particular sequence in genome where DNA replication begins. May be single(prokaryotes) or multiple(eukaryotes).
|
|
|
DNA Replication -
Replication Fork |
Y-shaped region along DNA template where leading and lagging strands are synthesized.
|
|
|
DNA Replication -
Helicase |
Unwinds DNA at replication fork
|
|
|
DNA Replication -
Single Stranded Binding Proteins(SSBPs) |
Prevents strands from reannealing
|
|
|
DNA Replication -
DNA topoisomerases Relevant Medication/Disorder |
Create a nick in the helix to relieve supercoils created during replication
Note: Fluoroquinolones - Inhibit DNA gyrase(specific prokaryotic topoisomerase) |
|
|
DNA Replication -
Primase |
Makes an RNA primer on which DNA polymerase III can initiate replication
|
|
|
DNA Replication -
DNA Polymerase III |
Prokaryotic ONLY, Elongates leading strand by adding DNA to the 3' end. Elongates lagging strand until it reaches primer of preceding fragment. Has 3' to 5' exonuclease activity which "proofreads" each added nucleotide.
DNA Pol III - 5' to 3' synthesis and 3' to 5' proofread |
|
|
DNA Replication -
DNA Polymerase I |
Prokaryotic ONLY. Degrades RNA primer and fills in the gap with DNA.
DNA Pol I - DNA polymerase I excises RNA primer with 5' to 3' exonuclease. |
|
|
DNA Replication -
DNA Ligase |
Seals.
|
|
|
Describe DNA replication and all important enzymes.
|
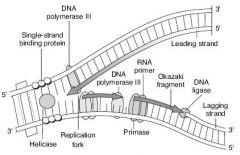
|
|
|
DNA Repair -
Nucleotide Excision Repair What type of damage does it repair? Relevant Medication/Disorder |
Specific endonucleases release the oligonucleotide containing damaged bases; DNA polymerase and ligase fill and reseal the gap, respectively.
Repairs damage that can occur to bases from a vast variety of sources including chemicals, radiation and other mutagens. These repairs tend to be bulky helix-distorting lesions, including UV-induced thymine dimers and 6'-4' photoproducts. Mutated in Xeroderma Pigmentosum(dry skin with melanoma and skin cancers, "children of the night"), which prevents repair of thymidine dimers |
|
|
DNA Repair -
Base Excision Repair What type of damage does it repair? Relevant Medication/Disorder |
Specific glycosylases recognize and remove damages bases, AP endonuclease cuts DNA at apyrimidinic site, empty sugar is removed, and the gap is filled and resealed.
It is responsible primarily for removing small, non-helix-distorting base lesions from the genome such as oxidized, alkylated or deaminated bases. |
|
|
DNA Repair -
Mismatch Repair Relevant Medication/Disorder |
Unmethylated, newly synthesized string is recognized, mismatched nucleotides are removed, and the gap is filled and resealed.
Mutated in Hereditary Nonpolyposis Colorectal Cancer(HNPCC) |
|
|
DNA Repair -
Nonhomologous end joining |
Brings together 2 ends of DNA fragments. No requirement for homology.
|
|
|
DNA/RNA/protein synthesis direction
|
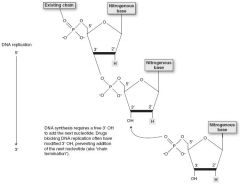
DNA and RNA are both synthesized 5' to 3'. Remember that the 5' of the incoming nucleotide bears the triphosphate(energy source for the bond). The 3' hydroxyl of the nascent chain is the target.
mRNA is read 5' to 3'. Protein synthesis is N to C. |
|
|
Types of RNA
|
rRNA - most abundant type of RNA
mRNA - longest type of RNA tRNA - smallest type of RNA Rampant, Massive, Tiny |
|
|
Start and Stop Codons -
mRNA Start Codon Eukaryotes vs Prokaryotes |
AUG(or rarely GUG)
Eukaryotes - Codes for methionine, which may be removed before translation is completed. Prokaryotes - Codes for formyl-methionine(f-Met) AUG inAUGurates protein synthesis. |
|
|
Start and Stop Codons -
mRNA Stop Codon |
UGA, UAA, UAG
U Go Away U Are Away U Are Gone |
|
|
Functional Organization of the Gene
|
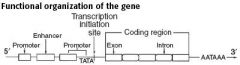
|
|
|
Regulation of Gene Expression -
Promoter Relevant Medication/Disorder |
Site where RNA polymerase and multiple other transcription factors bind to DNA upstream from gene locus(AT-rich upstream sequence with TATA and CAAT boxes)
Promoter mutation commonly results in dramatic decrease in amount of gene transcribed.. |
|
|
Regulation of Gene Expression -
Enhancer |
Stretch of DNA that alters gene expression by binding transcription factors.
Enhancers and silencers may be located close to, far from, or even within(in an intron) the gene whose expression it regulates. |
|
|
Regulation of Gene Expression -
Silencer |
Site where negative regulators(repressors) bind.
Enhancers and silencers may be located close to, far from, or even within(in an intron) the gene whose expression it regulates. |
|
|
RNA Polymerases -
Eukaryotes Which polymerase opens DNA? Where? |
RNA Polymerase I makes rRNA
RNA Polymerase II makes mRNA RNA Polymerase III makes tRNA None of them have a proofreading function but they can initiate chains. RNA Polymerase II opens DNA at promoter site. |
|
|
RNA Polymerase -
Prokaryotes Relevant medication/disorder. |
1 RNA Polymerase(multi-subunit complex) makes all three kinds of RNA.
α-amanitin(found in death cap mushrooms) inhibits RNA polymerase II. Causes liver failure if ingested. |
|
|
RNA Processing -
|
Occurs in nucleus(therefore only eukaryotes). After transcription:
1.) Capping on 5' end(7-methylguanosine) 2.) Polyadenylation on 3' end(about 200 A's) 3.) Splicing out introns initial transcript is called heterogeneous nuclear RNA(hnRNA). Capped and tailed transcript is called mRNA. ---ONLY processed RNA is transported outside the nucleus. ---AAUAAA = polyadenylation signal. ---Poly-A polymerase does not require a template. |
|
|
Splicing of pre-mRNA
|
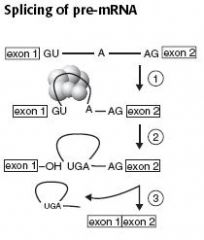
pre-mRNA splicing occurs in eukaryotes.
1 - Primary transcript combines with single nucleotide ribonuclear proteins(snRNP) and other proteins to form spliceosome. 2 - Lariat-shaped(looped) intermediate is generated. 3 - Lariat is released to remove intron precisely and join 2 exons. Patients with lupus make antibodies to spliceosomal snRNPs. |
|
|
Introns vs Exons
Relevant medication/disorders. |
Exons contain the genetic information coding for protein. Introns are intervening non-coding segments of DNA.
INtrons are INtragenic. Between genes. EXons are EXpressed. Notably, different exons can be expressed by alternative splicing to make unique proteins in different tissues. Example: ß-thalassemia mutations. |
|
|
tRNA Structure
|
75-70 nucleotides, 2" structure, cloverleaf form, anticodon end is opposite 3' aminoacyl end. All tRNAs, both eukaryotic and prokaryotic, have CCAat 3' end along with a high percentage of chemically modified bases. The amino acid is covalently bound to the 3' end of the tRNA.
|
|
|
tRNA Charging
Relevant medication/disorder |
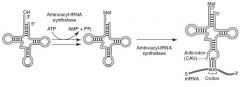
Aminoacyl-tRNA synthetase(1 per AA, "matchmaker", uses ATP) scrutinizes AA before and after it binds to tRNA. If incorrect, bond is hydrolyzed. The AA-tRNA bond has energy for formation of peptide bond.
A mischarged tRNA reads usual codon but inserts wrong amino acid. Aminoacyl-tRNA synthetase and binding of charged tRNA to codon are responsible for the accuracy of AA selection. Tetracyclines binds 30S subunit, preventing attachment of aminoacyl-tRNA |
|
|
Protein Synthesis -
Elongation Relevant medication/disorder |

1.) Aminoacyl-tRNA binds to A site(except for initiator methionine)
2.) ribosomal rRNA(aka "ribozyme") catalyzes peptide bond formation, transfers growing polypeptide to amino acid in A site 3.) Ribosome advances 3 nucleotides toward 3' end of RNA, moving peptidyl RNA to P site(translocation) Chloramphenicol inhibits 50S peptidyltransferase Macrolides bind 50S, blocking translocation Clindamycin binds 50S blocking translocation |
|
|
Protein Synthesis -
Termination Relevant medication/disorder |

Completed Protein is released from ribosome through simple hydrolysis and dissociates
Aminoglycosides, chloramphenicol, macrolides and clindamycin all act as protein synthesis inhibitors. |
|
|
Protein Synthesis -
Initiation Ribosomal Sizes Type of Energy Used Relevant medication/disorder |

Activated by GTP hydrolysis, initiation factors(eIFs) help assemble the 40S ribosomal subunit with the initiator tRNA and are released when the mRNA and the ribosomal subunit assemble with the complex.
Eukaryotes: 40S + 60S --> 80S Even Prokaryotes: 30S + 50S --> 70S Odd ATP - tRNA Activation (charging) GTP - tRNA Gripping and Going places (translocation) Aminoglycosides inhibit formation of the initiation complex and cause misreading of mRNA |
|
|
Energy requirements of translation
|
tRNA aminoacylation ATP --> AMP
Loading tRNA into ribosome GTP --> GDP Translocation GTP --> GDP Total Energy Expenditure: 4 high energy phosphoanhydride bonds. |
|
|
Posttranslational modifications
|
Trimming - Removal of N- or C-terminal propeptides from zymogens to generate mature proteins.
Covalent alterations - Phosphorylation, glycosylation, and hydroxylation. Proteosomal Degradation - Attachment of ubiquitin to defective proteins to tag them for breakdown. |
|
|
Cell Cycle Phases
|
Checkpoint control transitions between phases of cell cycle. This process is regulated by cyclins, CDKs, and tumor supressors. Mitosis(shortest phase): prophase-metaphase-anaphase-telophase.
G1 and G0 are of variable duration. |
|
|
Regulation of Cell Cycle -
CDKs |
cyclin-dependent kinases; constitutive and inactive
|
|
|
Regulation of Cell Cycle -
Cyclins |
Regulatory proteins that control cell cycle events; phase specific; activate CDKs.
|
|
|
Regulation of Cell Cycle -
Cyclin-CDK complexes |
Must both be activated and inactivated for cell cycle to progress.
|
|
|
Regulation of Cell Cycle -
Tumor Supressors |
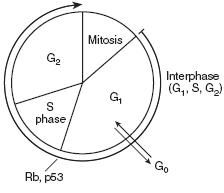
Rb and p53 normally inhibit G1-to-S progression; mutations in these genes result in unrestrained growth.
|
|
|
Cell Types -
Permanent |
Remain in G0, regenerate from stem cells
Example: neurons, skeletal muscle, cardiac muscle, RBCs |
|
|
Cell Types -
Stable(Quiescent) |
Enter G1 from G0 when stimulated
Example: hepatocytes, lymphocytes |
|
|
Cell Types -
Labile |
Never go to G0, divide rapidly with a short G1
Example: bone marrow, gut epithelium, skin, hair follicles |
|
|
Rough Endoplasmic Reticulum(RER)
RER in neurons What cells rich in RER? |
Site of synthesis of excretory proteins(exported) and of N-linked oligosaccharide addition to many proteins.
Nissl Bodies(RER in neurons) synthesize enzymes(e.g. ChAT) and peptide neurotransmitters. Mucus-secreting goblet cells of the small intestine and antibody-secreting plasma cells are rich in RER. |
|
|
Smooth Endoplasmic Reticulum(SER)
Which cells rich in SER? |
Site of steroid synthesis and detoxification of drugs and poisons.
Liver hepatocytes and steroid hormone-producing cells of the adrenal cortex are rich in SER. |
|
|
Golgi Apparatus
|
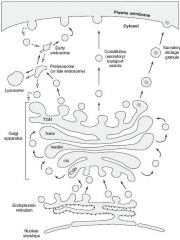
1.) Distribution center of proteins and lipids from ER to the plasma membrane, lysosomes, and secretory vesicles.
2.) Modifies N-oligosaccharides on asparagine 3.) Adds O-oligosaccharides to serine and theronine residues. 4.) Addition of mannose-6-phosphate to specific lysosomal proteins ---> targets the protein to the lysosome 5.) Proteoglycan assembly from core proteins 6.) Sulfation of sugars in proteoglycans and of selected tyrosine on proteins. |
|
|
Vesicular Trafficking Proteins in Golgi Apparatus
|
COPI - retrograde. Golgi --> ER
COPII - anterograde. RER --> cis-Golgi Clathrin - trans-Golgi --> lysosomes, plasma membrane --> endosomes(receptor mediated endocytosis) |
|
|
I-cell disease(Inclusion Cell Disease)
|
inherited lysosomal storage disorder; failure of addition of mannose-6-phosphate to lysosome proteins(enzymes are secreted outside the cell instead of being targeted to the lysosome). Results in coarse facial features, clouded corneas, restricted joint movement, and high plasma levels of lysosomal enzymes. Often fatal in childhood.
|
|
|
Microtubule
Molecular Motor Proteins Relevant medication/disorder |
Cylindrical structure composed of a helical array of polymerized dimers of α- and ß-tubulin. Each dimer has 2 GTP bound. Incorporated into flagella, cilia, mitotic spindles. Grows slowly, collapses quickly. Also involved in slow axoplasmic transport in neurons.
Molecular Motor Proteins - transport cellular cargo toward opposite ends of microtubule tracks. Dynein = retrograde to microtubule(+ to -) Kinesin = anterograde to microtubule(- to +) Chediak-Higashi syndrome - microtubule polymerization defect resulting in decreased phagocytosis. Results in recurrent pyogenic infections, partial albinism, and peripheral neuropathy. |
|
|
Microtubule Acting Drugs
|
Medendazole/thiabendazole - antihelminthic
Griseofulvin - antifungal Vincristine/Vinblastine - anti-cancer Paclitaxel - anti-breast cancer Colchicine - anti-gout |
|
|
Cilia Structure
Relevant medication/disorder |
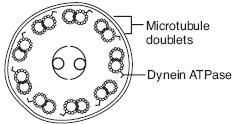
9+2 arrangement of microtubules
Axonemal Dynein - ATPase that links peripheral 9 doublets and causes bending of cilium by differential sliding of doublets Kartagener's Syndrome - immotile cilia due to a dynein arm defect. Results in male and female infertility(sperm immotile), bronchiectasis, and recurrent sinusitis(bacteria and particles not pushed out); associated with situs inversus. |
|
|
Cytoskeletal Elements -
Actin and Myosin |
Actin and Myosin - Microvilli, muscle contraction, cytokinesis, adherens junctions
|
|
|
Cytoskeletal Elements -
Microtubules |
Microtubule - Cilia, flagella, mitotic spindle, neurons, and centrioles
|
|
|
Cytoskeletal Elements -
Intermediate Filaments |
Intermediate Filaments - Vimentin, Desmin, cytokeratin, glial fibrillary acid proteins(GFAP), neurofilaments
|
|
|
Plasma Membrane Composition
What does high cholesterol cause? |
Asymmetric Fluid Bilayer
Contains choleterol(~50%), sphingolipids, glycolipids and proteins. High cholesterol or long saturated fatty acid content --> increased melting temperature, increased fluidity |
|
|
Immunohistochemical Stains -
Vimentin |
Connective Tissue
|
|
|
Immunohistochemical Stains -
Desmin |
Muscle
|
|
|
Immunohistochemical Stains -
Cytokeratin |
Epithelial Cells
|
|
|
Immunohistochemical Stains -
GFAP |
Neuroglia
|
|
|
Immunohistochemical Stains -
Neurofilaments |
Neurons
|
|
|
Sodium Pump
Relevant medication/disorder |
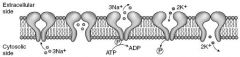
Na+-K+ ATPase is located in the plasma membrane with the ATP site on cytoplasmic side. For each ATP consumed, 3 Na+ go out and 2 K+ come in. During cycle, pump is phosphorylated.
Ouabain - inhibits ATPase by binding to K+ site Cardiac Glycosides(digoxin and digitoxin) directly inhibit the Na+-K+ ATPase, which leads to indirect inhibition of Na+/Ca+2 exchange. This increase [Ca+2] intracellularly, which increases cardiac contractility. |
|
|
Collagen
|
Most abundant protein in the human body. Extensively modified. Organizes and strengthens the extracellular matrix.
Type I(90%) - Bone, Skin, Tendon, dentin, fascia, cornea, late wound repair. |

