![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
133 Cards in this Set
- Front
- Back
|
What is meant by DNA having "polarity"?
|
5' PO3- and 3' OH groups
|
|
|
a. How is the backbone of DNA formed?
b. What makes DNA different from RNA? c. Where are the bases attached to on the Deoxyribose? |
a. 3'-5' Phosphodiester bonds
b. Missing OH group in the 2' position on the ribose in DNA c. Bases are attached at the C1 position on the ribose ring |
|
|
Why are GC bonds stronger than AT?
What are the implications for high GC areas? |
GC has 3 H-bonds, while AT only has 2
High GC areas are harder to break apart. Higher Tm |
|
|
What does hyperchromicity refer to when examining DNA at a wavelength of 260?
|
It means that there is denaturation of the DNA and increased absorbance
|
|
|
What is the most common Chromosomal form of DNA?
Is it right or left handed? How many base pairs per turn? |
B-DNA
Right Handed 10 bp per turn |
|
|
What is A-DNA?
How many BP per turn? Right handed or Left handed? |
Dehydrated form of B-DNA
11bp per turn Right Handed |
|
|
When is Z-DNA present?
How many BP per turn? Left handed or Right Handed? |
Present when there is a high GC content
May be induced during supercoiling to allow for transcription 12 bp per turn Left Handed |
|
|
a. What are Double Minutes?
b. Where are they found? c. What do they contain? |
a. Small fragments of extrachromosomal DNA
b. Found in Tumors: breast, lung, ovary, colon, neuroblastoma c. Contain oncogenes |
|
|
a. What are examples of Structural RNA?
b. What are examples of Regulatory RNA? |
a. tRNA, rRNA, 5SRNA
b1. miRNA, RNAi (micro-RNA, interfaceRNA - silences expression) b2. lncRNA (long, non-coding RNA. Regulation of expression) b3. Antisense RNA - blocks expression by binding mRNA |
|
|
What percentage of the Human Genome is protein-coding genes?
|
3%
|
|
|
What base pair combo is found most in Gene Dense areas?
What base pair combo is found most in Gene Poor Areas? |
Gene Dense - GC
Gene Poor - AT |
|
|
a. What percentage of the Eukaryotic genome is Tandem Repeats vs. Interspersed Repeats?
b. Where are Tandem Repeats found? |
a1. Tandem - 10-15% (highly repetitive)
a2. Interspersed - 25-40% (moderately repetitive) b. Tandem Repeats are found in Centromeric and Telomeric DNA |
|
|
What are SINEs and LINEs?
|
SINEs - Short Interspersed Nuclear Elements - Mostly Alu repeat sequences, which contain a recognition site for Alu1, a restriction enzyme
LINEs - Long Interspersed Nuclear Elements - Flanked by Alu sequences |
|
|
In Mitochondrial DNA, how many genes code for the ETC?
|
13
|
|
|
Why are mitochondrial DNA only passed on from the women?
|
Mitochondria from sperm are proteolytically degraded by autophagosomes
|
|
|
Mitochondrial DNA and Diseases
a. What are the most affected systems/organs? b. What are the symptoms of mutation of mitochondrial DNA? |
a. CNS, Muscle (myopathy), Kidney, Liver
b1. Increased Pyruvate --> Lactic Acidosis b2. Decrease in ETC b3. Increased [FA] --> High TG |
|
|
Leber's Hereditary Optic Neuropathy (LHON)
a. Symptoms b. Causes |
a. Sudden blindness and CNS disturbances
b. Due to point mutation in genes for Complex 1 of ETC |
|
|
Myoclonic Epilepsy & Ragged Red Fiber Disease (MERRF)
Symptoms: Causes: |
a. Muscle spasms and ataxia
b. Due to point mutation in Lys on tRNA |
|
|
a. What is DNA methylation used for?
b. What enzyme carries out methylation? c1. At which sites do methylations occur? c2. Why does methylation occur there? |
a. Regulation of gene expression
b. Methyltransferases (Dnmts) c1. Almost exclusively at CpG (C-G) sites c2. CpG Islands precede genes in or near promotors. If methylated, there will be no gene expression |
|
|
a. What is a nucleosome?
b. What is a Histone? |
a. DNA associated with a histone
b. Histone - highly basic and polycationic protein where Arg & Lys(>20%) |
|
|
a. What is the purpose of Histone Acetylation and Deacetylation?
b. What enzymes perform Histone Acetylation and Deacetylation? |
a. To unwind or tighten, respectively, the interactions between the histone and DNA
b1. Histone acetyltransferases(HAT) Transfers Acetyl group from Acetyl-CoA --> Lys on Histone b2. Histone deacetylase (HDAC) Restores the positive chargetighter DNA/Histone intx |
|
|
a. What is Say-Barber-Biesecker-Young-Simpson syndrome (SBBYSS or Ohdo syndrome)?
b. What is its cause? |
a. A multiple anomaly syndrome characterized by severe intellectual disability, blepharophimosis(eyelid malformation) and a mask-like facial appearance
b. Downregulation of KAT6B gene, which codes for Histone Acetyl Transferase (HAT). |
|
|
a. Where, in relation to the replication fork, is positive supercoiling of DNA? Where is negative supercoiling?
b. Does most DNA have positive or negative supercoiling? |
a. Positive (overtwisting): ahead of the replication fork
Negative (undertwisting): Behind the replication fork b. Most DNA has NEGATIVE supercoiling |
|
|
a. What is the function of Topoisomerases?
b. What are the actions and energy requirements of Topoisomerase I and II? |
a. Relief of supercoiling
b. Topo I: Cuts a single strand of DNA - No ATP required Topo II: Cuts both strands of DNA - ATP required |
|
|
What is the source of the nucleotide that is added to the 3' OH end of DNA in DNA synthesis? What is added and what is released?
|
The Alpha Phosphate of Nucleotide Triphosphate (NTP) is added and PPi is released
|
|
|
a. In what direction does DNA synthesis happen?
b. In what direction does proofreading happen? |
a. 5' --> 3'
b. 3' --> 5' |
|
|
What are the steps in the formation of the replication fork in DNA synthesis?
|
1. DNA Helicase unwinds the DNA helix
2. TopoII (gyrase) relieves the stress by cutting/unwinding 3. Single Stranded Binding Proteins bind to the DNA to prevent reannealing 4. DNA Pol III elongates the leading strand from an RNA primer 5. Primase moves toward the replication fork on the lagging strand to lay down primers 6. DNA Pol III extends the RNA primers w/ DNA bases to make Okazaki fragments 7. Pol I removes RNA primers and fills in DNA bases 8. DNA ligast connects the phosphodiester backbone (ATP --> AMP + PPi) |
|
|
How does proofreading happen?
|
If polymerase mispairs a nucleotide with the temeplate, it uses its 3'-5' exonuclease activity to excise the mismatched nucleotide
|
|
|
What is homologous recombination?
|
A form of DRBR (double stranded break repair) in which a homologous sister chromatid is sought out to use as a template for the repair of the broken DNA strands.
|
|
|
a. What is DSBR (double stranded break repair) called when it happens at a replication fork?
b. What does this process use in order to repair the nick? |
a. Break-Induced Replication (BIR)
b. Homologous Recombination (HR) |
|
|
a. What does Etoposide do?
b. What is Etoposide used for? |
a. Targets Eukaryotic TopoII and binds the TopoII-DNA complex, causing DNA double strand breaks --> Cell Death
b. Used to treat lung and testicular cancer, lymphoma, non-lymphocytic leukemia, and glioblastoma multiforme (brain tumor) |
|
|
a. What are examples of quinolones?
b. What do they do? c. What are they used for? |
a. Ciprofloxacin and Nalidixic Acid
b. Targets bacterial DNA gyrase (Top II) for inhibition c. Treats UTI and Lower Respiratory Infections |
|
|
a. What is a telomere and what does it consist of?
b. What maintains the telomeres? c. How does this enzyme work? d. How do we combat this enzyme in cancer cells? |
a. The ends of chromosomes complexed with proteins. They consist of repeats of AG3T2 (AGGGTT), which forms a T-loop for stability
b. Telomerase (only in germ cells and cancer cells - NOT in somatic cells) c1. Telomerase uses Reverse Transcriptase (hTERT) to extend the longer DNA strand further, using RNA (hTERC) as the template to make DNA c2. Primase lays an RNA primer on the short strand and Polymerase completes the strand. c3. The short end now is complete, but the long end has been extended. That "overhang" forms the T-loop for protection from nucleases d. Imetelstat - leads to progressive shortening of telomeres and reduced rates of proliferation. Eventually, this leads to cell death. |
|
|
a. What is Dyskeratosis Congenita?
b. What is the symptomology and prognosis? |
a. A disease in which telomeres get short much more quickly than normal
b1. This leads to premature aging and death. b2. Higher risk of life-threatening infections, leukemia and other blood cancers, intestinal disorders, cirrhosis of the liver and pulmonary fibrosis, and deadly stiffening of lung tissue |
|
|
What are the main differences b/w Prokaryote and Eukaryote DNA synthesis?
|
1. Prokaryotes can do it throughout the cell cycle, but Eukaryotes can only do it during S-Phase
2. Prokaryotic DNA is much smaller and can replicate faster as a result 3. Eukaryotic cells have more polymerase molecules to compensate for slower replication fork 4. Prokaryotes have ONE origin of replication (Ori C), while Eukaryotes have multiple Autonomous Replication Sites (ARS) simultaneously (replication bubbles) |
|
|
a. What controls the cell cycle?
b. How is the cell cycle controlled? c. What happens if RB gene for Rb protein is mutated? |
a. Cyclins
b1. Cyclins help in maneuvering from one part of the cell cycle to another b2. Cyclin Dependent Kinases (CdK) cause phosphorylation of proteins such as the Retnoblastoma protein (Rb), which is central in cell cycle regulation b3. If dephosphorylated, it binds E2F (transcription factor) and repressed transcription. If phosphorylated, it cannot bind E2F, so transcription is activated b4. Rb-phosphorylated in mid to late G1 by cyclin/CdK Complex, so transcription is activated c. It is a tumor suppressor gene, so Retinal Malignancy can develop. Also carcinomas of the bladder, lung, liver, and prostate. |
|
|
What are the most common causes of DNA damage?
|
1. OXIDATIVE DAMAGE IS THE MOST COMMON FORM - Cellular Metabolism - free radicals are often created b/c oxygen is involved
2. UV Light Exposure - Creates a Thymine Dimer, a covalent bond b/w adjacent T-T, which can block DNA replication 3. Ionizing Radiation - Due to ionization , there can be pairing b/w mismatched bases such as T-G 4. Chemical Exposure - Nitrous Acid, formed from digestion of nitrites used as preservatives in foods, change C --> U. Nitrogen mustards will act as alkalating agents as well, although we can use this in chemotherapy. 5. Replication Errors |
|
|
a. What are the main DNA repair mechanisms?
b. How do they work? c. When do they happen? |
a1. Nucleotide Excision Repair - Spontaneous, chemical, or radiation damage to a DNA segment - Fixed by removal of up to 30 nucleotides and replacement
a2. Mismatch Repair (MM) - Copying errors fixed by methyl-directed strand cutting, exonuclease digestion, and replacement a3. Base Excision Repair (BER) - For Spontaneous, Chemical, Oxidative, or Radiation Damage - Base removal by N-glycosylase, abasic sugar removal, replacement c. You want the repair to happen BEFORE the next round of replication b/c you don't want the mutation becoming permanent |
|
|
a. How is HNPCC (Hereditary Nonpolyposis Colon Cancer) associated with defective DNA repair?
b. How is XP (Xeroderma Pigmentosum) associated with defective DNA repair? c. How is Breast Cancer associated with defective DNA repair? |
a. Defect in MMR (hMSH1 and 2 mutations)
b. Defect in NER - No removal of Thymine Dimers, so sensitivity to UV light. Large increase in skin cancer risk c. Defect in Homologous Repair for DSBR (Mutations in BRCA1 and BRCA2, which are tumor suppressors). Also a marker for prostate cancer risk in men. |
|
|
In DNA base changes, what are Transitions and Transversions?
|
Transitions: Purine changes to another purine or Pyrimidine changes to another Pyrimidine
Transversions: Purines become Pyrimidines and vice versa |
|
|
What is the process to fix DNA when an incorrect base is inserted? (Prokaryote)
|
Methyl-Directed Mismatch Repair (MMR)
1. Mut proteins ID the G(A-methyl)TC sequences on parent strand. 2. Endonuclease Cuts Strand 3. Exonuclease removes bases 4. Gap is filled by DNA Pol 5. DNA Ligase joins the bases |
|
|
What is the process to fix DNA when a single base is mutated such as C --> U?
|
Base Excision Repair (BER)
1. A single base is removed 2. Phosphodiester backbone is nicked by endonuclease 3. Single empty sugar-P residue is removed by Lyase 4. Gap is filled by DNA(delta) using sister strand as template 5. Phosphodiester backbone is closed by Ligase |
|
|
What is the process to fix Thymine dimers (caused by UV light) in DNA?
|
Nucleodite Excision Repair (NER)
1. uvrABC excinuclease (UV damage specific endonuclease) recognizes dimers and cleaves the damaged strand 5'-3' 2. Gap is filled in by DNA Polymerase 3. Bases are joined by DNA Ligase |
|
|
What are the steps in elongation?
|
DNA Pol III adds NTP to 3' OH
DNA Ligase releases PPi |
|
|
1. What are the functions of Pol I?
2. What are the functions of Pol II? 3. What are the functions of Pol III? What makes processivity so effective? |
1a. 5'-->3' Exonuclease - Removes RNA primer and proofs
1b. 5'-->3' Polymerase - Replace RNA w/ DNA 1c. 3'-->5' Exonuclease - Proofreading 2. 5'-->3' Polymerase - Repair 3a. 3'-->5' Exonuclease - Proofreading 3b. 5'-->3' Polymerase - Replication of BOTH strands 3c. Beta subunit clamping makes for high processivity |
|
|
What is the process of BIR? (break induced repair)
Can crossover occur? |
Uses Homologous Recombination
Forms Holliday Junctions using Recombinase for strand invasion Resolvases resolve the Holliday Junctions Crossover can occur |
|
|
What does Cyclophosphamide treat?
What causes the initial mutation? |
Tumors associated with Chemical change
C-->U mutation due to Nitrites (preservatives in food) OR alkylating agents (N-Mustards) |
|
|
What is 8-O2-deoxyG?
How do you get this? What kind of damage is this? What else causes this kind of damage? What other diseases are associated with it? |
Modified RNA base and ROS
From smoking and asbestos Oxidative damage Inflammation, aging, smoking Alzheimer's, Atherosclerosis, Diabetes |
|
|
What is used as a template in most DNA repaie mechanisms?
|
The undamaged strand
|
|
|
What is the process to fix DNA when an incorrect base is inserted? (Eukaryote)
|
MMR - Directed by a 5' Strand Break
1. MutS(alpha) Protein activates EXO1 2. EXO1 hydrolyzes the incised strand from the 5' end 3. DNA(delta) puts correct DNA in gap 4. Ligase connects the gap |
|
|
What are the 2 kinds of DSBR?
|
1. NHEJ - non-homologous end joining
A stop-gap measure to bind exposed strands to each other 2. Homologous Recombination (HR) Uses Homologous sister chromatid as a template. Uses RAD Proteins and BRCA Proteins (strand invasion) |
|
|
Does RNA:
1. Have hairpin loops? 2. Require a primer? 3. Have a 2'OH on Ribose? |
Yes
No Yes |
|
|
What is a Cis-natural antisense transcript?
What is it used for? |
Transcript made from the opposite strand of DNA at the same location
Used to block mRNA from making protein |
|
|
Does Eukaryotic mRNA:
Have a repair system? Have 5' and 3' UTRs? Have a long half life? Hairpin loop secondary structure? |
No
Yes Yes Yes |
|
|
Why is the TATA box used to start transcription?
|
It is easy to pull AT apart
|
|
|
1. What are the sizes of the ribosomes in Eukaryotic rRNA?
2. What are the sizes of the ribosomes in Prokaryotic rRNA? |
1. Eukaryote - 80S ribosome made of 60S (28S has ribozyme activity) and 40S
2. Prokaryote - 70S made of 50S (23S has ribozyme activity) and 30S |
|
|
In what direction foes tRNA read the codon?
What is the structure of tRNA? How many species of tRNA are there for how many AA? |
5'-->3'
Cloverleaf 61 species for 20 AAs |
|
|
What are the functions of lncRNA?
|
1. Signaling
2. Decoys - titrate away protein factors 3. Guides - Localize ribonucleoprotein complexes to targets - ALTERS GENE EXPRESSION 4. Scaffolds - links protein complexes 5. X-Chromosome Inactivation - Methylates DNA and histones |
|
|
What are the subunits of Prokaryotic RNA Polymerase?
What is a holoenzyme? What drug works against it and for what disease? How is reading and encoding done? |
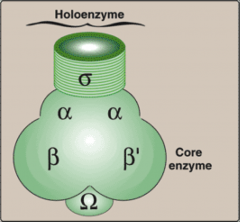
alpha, alpha', beta, beta', omega
Sigma Subunit - Transcription initiation factor - Recognizes the promoter Rifampin - Inhibits the Beta Subunit of RNAP - Used for TB Reading 3'-->5' and coding 5'-->3' |
|
|
In Prokaryotic DNA:
1. What is the non-template/sense/coding strand? 2. What is the template/nonsense/non-coding strand? |
1. Same sequence as mRNA, but with T instead of U
2. Opposite sequence as mRNA so that mRNA will have the same code as the sense strand |
|
|
What are Rho dependent and Rho independent mechanisms of termination? (Prokaryotic uses Rho)
|
Independent - Hairpin Loop -->AU bonds break --> PolyU tail pulls out of Pol active site
Dependent - Rho recognizes mRNA, displaces DNA strand, and the mRNA and DNA dissociate |
|
|
What kind of RNA does RNA Pol I, Pol II, and Pol III make in Eukaryotes?
|
Pol I - rRNA
Pol II - mRNA Pol III - tRNA, 5SRNA, snRNA |
|
|
What does alpha Amanitin do and where does it come from?
What are the consequences? |
Comes from Poison Mushroom. Binds RNA Pol II
Consequence - You make NO mRNA --> No proteins made |
|
|
What are the positions of the Promotors and enhancers?
|
CAAT Box is -85 upstream
TATA Box is -25 upstream |
|
|
What do Transcription Factors bind to?
|
DNA elements
|
|
|
How do enhancers interact with promotors from so far away?
|
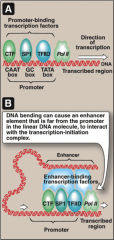
Enhancers bend DNA when they bind to enhancer elements and interact with the TFs
|
|
|
What is the process for termination in Eukaryotes?
|
Pol II transcribes past the PolyA site, stops, and then cleaves.
Upstream cleavage product is PolyAdenylated by Polyadenylate Polymerase and downstream product is degraded |
|
|
What is the most conserved gene in all cells?
|
rRNA
|
|
|
What sequences are never transcribed?
|
Promoter and Enhancer regions
|
|
|
Where on a tRNA does the charged AA bind?
What enzyme adds the AA? |
The CCA sequence on the 3' end
Nucleotidyltransferase |
|
|
What is the primary purpose of the Nucleolus?
How is a ribosome assembled? |
Makes rRNA
Proteins imported from the cytoplasm, form subunits in the nucleolus, exported back to the cytoplasm They then synthesize proteins in the cyto |
|
|
How is the 5'-7methylG cap made?
What is its function? |
1. Nucleus - Guanylyltransferase adds the cap
2. Cytosol - Methylation Function 1. Protects mRNA from degradation 2. Increases translatability 3. Enhances transport to cytoplasm 4. Enhances efficiency of splicing mRNA 5. Recognized by Initiation factor eIF4e |
|
|
How is the PolyA tail made?
What is the function of PABP (PolyA Binding Protein) What is the function of the PolyA tail? |
1. 200 Adenines are made after the AAUAAA signal
2. Done w/o a template 3. PolyA Binding Protein protects the tail Function 1. Stabilize mRNA 2. Facilitate exit from nucleus 3. Extends life of mRNA for multi-translation |
|
|
What mRNAs have no PolyA tail?
|
Histone and Heat Shock mRNA
|
|
|
What is the process for RNA intron splicing?
|
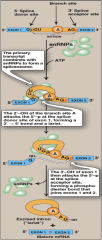
1. snRNP combines with the mRNA using ATP - Forms SPLICEOSOME
2. 2'OH from the branch site "A" attacks 5'P donor site from Exon 1 3. Forms Lariat 4. 3'OH from donor attacks the 5'P acceptor site 5. snRNP dissociate 6. Excised intron floats away, attached to itself |
|
|
What are the beginning and ending NTs in an Intron?
|
Beginning - GU
Ending - AG Starts and Ends with G (GUAG) |
|
|
What percentage of all genetic diseases are due to splicing mutations?
|
15%
|
|
|
What causes Beta-Thalassemia?
|
Point mutation creates TTAG before the end of the intron.
AG signals the end of an intron, so it is like a premature STOP codon mRNA is spliced and intron genes that were not removed get coded |
|
|
What is the cause of Systemic Lupus Erythromatosus (SLE)?
What are the symptoms? |
Autoimmune Disease
Antinuclear Antibodies (ANA) to dsDNA and U1RNP of Spliceosome Symptoms: Arthralgia, Butterfly rash on face and Pericarditis |
|
|
What is the ONLY Start Codon?
What AA does it code for? |
AUG - Methionine
|
|
|
What are the Stop Codons?
|
UAA, UAG, UGA
|
|
|
What is an Open Reading Frame (ORF)?
|
Sequence of Triplets that code for protein with a START and STOP codon
|
|
|
What is a missense mutation?
What are the two types of missense mutations and what do they do? |
Change in Nucleic Acid that may or may not affect the protein function
Conservative - AA changed to similar one Non-conservative - AA changed to entirely different one |
|
|
What is the cause of Sickle Cell Disease?
|
Position 6 Glu --> Val mutation due to A-->T missense mutation
|
|
|
What is a nonsense mutation?
What diseases are associated with it? |
Mutation that results in a STOP codon and truncated protein
Diseases: CF, Hemophilia, Retinitis Pigmentosa, Duchenne Muscular Dystrophy |
|
|
What are the 2 causes Cystic Fibrosis (CF)?
|
Nonsense mutation at NT 1609
Makes CAG (Gln) --> TAG (stop) Loses 1000 AA from protein Non-frameshift deletion of a triplet --> Loss of Phe Delta508 (90% of cases) The Ile that is next to it is not lost because of degeneracy |
|
|
What causes Duchenne's Muscular Dystrophy?
What is the therapy for this? What phenotype will the person have with this therapy? What drug might also help? |
Deletion - X-linked Recessive
Disrupts reading frame and leads to NONSENSE MUTATION downstream Absence of Dystrophin in Striated muscle Therapy: AON (antisense oligonucleotide) - Exon Skipping Skipping Exon 51 with a complementary mask will restore the reading frame Becker Phenotype - truncated protein, but less severe Gentamycin (aminoglycosides) and Ataluren allow ribosome to ignore premature stop codon (READTHROUGH) - AKA Nonsense Allele Inhibitor |
|
|
What are the types of Frame Shifts Mutations?
What disease is associated with Frame Shift Mutations? |
Base Insertion not divisible by 3 - Slippage of replicating strand necessitates extra nucleotide being added
Result - 1 with POS Frame shift and 1 Wild Type Strand Base Deletion - Slippage of parent strand results in loss of base not divisible by 3 Result - 1 with NEG Frame shift and 1 Wild Type Strand Ribosomal Slippage - Slippery Sequences of easily accessible codons. (Choke Pts are harder to access in Prokaryotes ONLY) Tay Sach's Disease (TSD) - 4bp insert into Exon 11 of HexA gene on Chromosome 15 Creates Premature STOP |
|
|
What is a deletion or insertion of a triplet called?
What disease is associated with it? |
Non-frameshift mutation
Cystic Fibrosis Non-frameshift deletion of a triplet --> Loss of Phe Delta508 (90% of cases) The Ile that is next to it is not lost because of degeneracy |
|
|
What is the most common TNR (trinucleotide repeat)?
What is the cause of TNRs? What is the danger of TNRs? What is "Anticipation"? |
CAG --> Gln (adds a string of Glutamine)
Causes - DNA Pol Slippage during replication Expansion of them with every Meiosis, so each generation will have MORE repeats. When it reaches a critical size, there will be impeded function Anticipation - Worstening of clinical phenotype with generations due to increased repeats |
|
|
How many TNR is considered normal?
What Diseases are associated with TNR? |
Less than 35
Huntington's - PolyQ disease Intracellular Aggregation of mutant protein Spinocerebellar Ataxia - PolyQ Myotonic Dystrophy - NON-PolyQ TNR of CTG (Leu); autosomal dominant |
|
|
What is the Wobble Hypothesis?
What does this hypothesis go against? |
5' end of an ANTICODON or 3' end of a CODON are at the wobble position
Degeneracy of code lets tRNA recognize multiple codons This goes against Watson Crick Base Pairing - Says there are exactly as many tRNA as codons for AA |
|
|
What is the direction of Translation?
|
5' (NH2) --> 3'(COOH)
|
|
|
How are AAs added to tRNA?
|
With an Ester Link to the 3' end
Uses ATP --> AMP + PPi (2 high energy bonds) |
|
|
What is the process of Translation in Prokaryotes?
|
1. Recognition of the Sine Delgarno Sequence - UAAGGAGG - Orients the ribosome
2. IF-2 binds GTP and finds the AUG (fMET) 3. Brings tRNAi to P site on 30S ribosome 4. Hydrolyze GTP 5. Another GTP for translocation (EF-G) 6. Another GTP for Elongation (EF-Tu) |
|
|
What is the rate limiting step in Translation (Eukaryotic)?
|
eIF-4 helicase orienting the Ribosome near the 5'CAP
|
|
|
What is the process of Translation in Eukaryotes?
|
1. 5'CAP recognition and then AUG START
2. eIF-2 binds GTP 3. Met-tRNAi comes into the P-site 4. GTP Hydrolyzed 5. Lage subunit binds - forms initiator complex 6. Elongation Factors bring new tRNA to A site (GTP hydrolyzed and EF recycled) 7. peptide bond formed via Peptidyl Transferase in Large Subunit (28S Ribozyme) 8. Translocation (EF-G) and GTP used 9. Termination uses GTP and release Factors |
|
|
How many ATP are used Per Peptide Bond in Translation?
|
4
|
|
|
What is the function of Aminoglycosides?
|
"mycins"
Binds 30S ribosome in Initiation of Translation Notes: Kills Kidneys |
|
|
What is the function of Tetracyclines?
|
"cyclins"
Blocks tRNA binding 70S ribosome in Translation Notes - Doxycycline - Lyme Disease |
|
|
What is the function of Chloramphenicol?
|
Clindamycin
Inhibits Peptidyltransferase by binding the 50S Ribosome in Translation Notes: Broad Spectrum |
|
|
What is the function of Macrolides?
|
Z-pack, Erythromycin
Binds 30S of the 70S ribosome and inhibits Translocation in Translation Notes: Gram (+) Cocci |
|
|
What is the function of Quinolones?
|
"fl"
Binds DNA Gyrase (TopoII) during Bacterial DNA Synthesis Notes: UTI, Food-bourne illness, Gram + Cocci |
|
|
What is the function of Rifamycin?
|
"Rifampin"
Binds to DNA dependent RNA Polymerase (Beta Subunit) during RNA Synthesis Notes: TB |
|
|
What is the function of Diphtheria Toxin?
|
Inactivates EF-2, blocking Translocation during Translation
|
|
|
What are the two types of Polysomes?
|
Free Ribosomes - No signal peptide. Proteins for Cyto, Mito, Nucleus, Peroxisomes
RER-bound Ribosomes - SIGNAL PEPTIDE, proteins for Membrans and Secretion |
|
|
Post Translational Modification - How does the signal peptide work?
|
1. 15-30 hydrophobic residues at the NH2 terminus
2. Needed for exporting proteins 3. Signal Recognition Particle binds signal 4. Brings Protein/Ribosome complex to ER emmbrane on SRP receptor 5. Translocator Pore Recognizes SP 6. Peptide thru translocator into ER membrane 7. SP Cleaved by Signal Peptidase 8. Completed peptide into ER 9. Mod and Export by Golgi |
|
|
What has a signal peptide?
|
Pre-pro-insulin
Pre-pro-collagen |
|
|
What removes Met or fMet from the N-terminus in Posttranslational modification?
|
Met-aminopeptidase
|
|
|
What is the Acetylation at the NH2 terminal called?
|
Degron - degradation signal
|
|
|
What is commonly Amidated at the C-terminal post-translationally?
|
Peptide hormones
|
|
|
What has Asn-N-linked Glycosylation as a post-translational Modification?
What has Asp-N-link as a post-translational mod? What is the link for Serine and Thr? |
Dolichol-Phosphate
Glycoproteins and Proteoglycans (Receptors, Matrix Proteins, Collagen) Thr or Ser O-linked |
|
|
What are the Posttranslational AA mods?
|
Phosphorylation: Thr, Tyr, Ser
Methylation: Arg and Lys (Positives) Hydroxylation: Pro and Lys Gamma Carboxylation: Glutamate - Vitamin K |
|
|
What disease is associated with deficiency in Hydroxylation of Proline and Lysine?
|
Ehler's Danlos
|
|
|
What 2 molecules require Disulfide bonds as a post-translational mod?
|
Insulin and Ribonuclease
|
|
|
What is the nucleophile on the Serine proteases and what is its function?
|
OH group on Seine --> Acivates Zymogens
|
|
|
How many AA long are Ubiquitin?
What is the size of a proteosome? What diseases are associated with the above things? |
76
26S Storage Disorders - Astrocytoma, Parkinson's, Alzheimer's |
|
|
What is the primary site of regulation of gene expression in Prokaryotes?
|
Transcription
|
|
|
What are constitutive genes in bacteria?
What are Regulated Genes and what is an example? |
Housekeeping genes - Continuously expressed
Genes expressed only when needed - Lac Operon |
|
|
Induceable genes code what kinds of enzymes?
When are genes induced? |
Induceable Enzymes
When their products are needed (catabolic enzymes) |
|
|
Repressable genes code for what kinds of enzymes?
When is transcription active? |
Repressable enzymes
Until products are no longer needed - Feedback inhibition (Anabolism) |
|
|
What proteins bind Operators?
|
Repressor Proteins
|
|
|
What are 2 regulatory mechanisms that work by controlling repressors?
|
Induction - Inducer INactivates repressor
Repression - Co-repressor required to activate repressor |
|
|
Where in DNA are regulator sequences located?
What does it mean when we say that sequences are "cis-acting"? What is means by "trans-acting"? Where are trans-acting proteins synthesized from? What kind of regulation do trans-acting elements do? |
Embedded in non-coding regions near genes
Cis-acting means same chromosome as the geen being regulated Trans-acting Elements (regulatory factors) bind cis-acting elements on DNA Different genes than the ones that they regulate Repression, Activation |
|
|
What proto-oncogenes are associated with AP-1 (activator protein 1)?
|
Jun and Fos
|
|
|
What are the 4 domains of Transcription Factors?
|
1. Activation Domain - Region that interacts w/ other proteins (such as Polymerase)
2. DNA Binding Domain - Recognizes specific regions near the start of transcription 3. Nuclear Localization Domain - Signal for TF to go to Nucleus after being synth. in cyto 4. Dimerization Domain - TF work as dimers |
|
|
What is Li-Fraumeni Syndrome?
|
When someone has only 1 functional copy of P53, they get tumors as an early adult
|
|
|
How is RNA Pol II Regulated?
What are the components of TFIIH? |
1. C-terminal Domain (CTD) is Phosphorylated by TFIIH
2. It becomes active, breaks away from the initiation complex, and Starts elongation TFIIH - Kinase, ATPase, Helicase |
|
|
What is the function of an Enhancer?
Does orientation or position matter? If there is no Promotor activity, what is the result? |
Binds proteins and increases promoter activity
Nope - bends the DNA Nothing - There is no protein made |
|
|
TF binding motifs - Zinc Finger
What kind of receptors have this? What binds the zinc? What is the structure? |
Steroid Hormones
Cys residues bind Zn2+ 2 antiparallel Beta Strands and 1 alpha-helix |
|
|
TF binding motifs - Leucine Zipper
How do the Leu associate with each other and create the secondary structure? Where on the DNA do the Basic AAs go? What are examples of Leu Zipper? What other motif is similar to the Leu Zipper? |
Hydrophobic Interactions
Major Groove c-fos, and c-jun Helix-Loop-Helix |
|
|
How do Statins work?
|
1. Block HMG-CoA Reductase --> Cholesterol synth in liver drops
2. SREBP to Golgi 3. Processed and sent to nucleus 4. SREBP binds SRE on DNA 5. Upregulation of LDL Receptor Gene and increase in LDL receptors 6. Blood LDL DROPS |
|
|
What are the Peptide Hormones?
How do Peptide Hormones work since they do not enter the nucleus? |
Epi, Insulin, Glucagon
1. They Activate cAMP 2. CRE (cAMP Response Elements) bind CREBP (CRE Binding Proteins) 3. cAMP leads to Phosphorylation of CREBP (via activated kinases) 4. Phosphorylation activates RNA Pol II and begins transcription |

