![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
70 Cards in this Set
- Front
- Back
|
Who was Friedrich Miescher and what did he discover?
|
A Swiss biologist who, in 1868, carried out chemical studies on a cell nuclei, detecting a phosphorous-containing nuclein, which had an acidic portion (DNA) and a basic protein (histone).
|
|
|
What was the Hershey-Chase experiment and what did it tell us?
|
Showed that DNA, but not protein, gives genetic information!
In 1952, Alfred Hershey and Martha Chase showed that the DNA of a bacteriophage T2, but not its protein coat, enters the host and provides genetic information for viral replication. |
|
|
What are nucleic acids and their components?
|
Nucleic acids (nitrogenous base + sugar) added to a phosphate group to make nucleotides!
Ex: Adenosine triphosphate Nitrogenous Base + Sugar + Phosphate |
|
|
Where do nucleic acids form bonds?
|
The nitrogenous base attaches at the 1'-Carbon of the sugar.
The Phosphate group attaches at the 5'-Carbon of the sugar. |
|
|
What is the difference between deoxyribose and ribose?
|
Deoxyribose has a Hydrogen on the 2'-Carbon and forms deoxyribonucleotides, which form deoxyribonucleic acids.
Ribose has an -OH group on the 2'-Carbon and form ribonucleotides, which form ribonucleic acids. |
|
|
What are the pyrimidine bases?
|
Cytosine
Thymine (DNA ONLY) Uracil (RNA ONLY) Big name, small structure. |
|
|
What are the Purine bases?
|
Adenine
Guanine Small name, big structure |
|
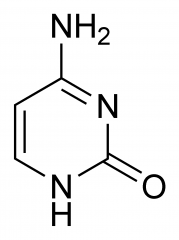
What nitrogenous base is this?
|
Cytosine
|
|
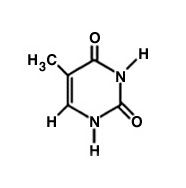
What nitrogenous base is this?
|
Thymine
|
|
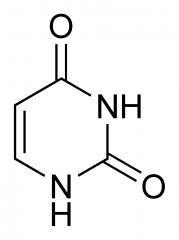
What nitrogenous base is this?
|
Uracil
|
|
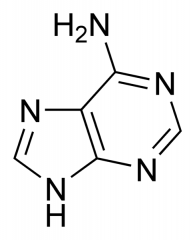
What nitrogenous base is this?
|
Adenine
|
|
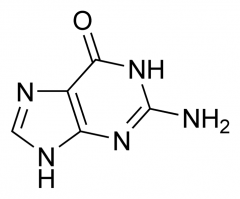
What nitrogenous base is this?
|
Guanine
|
|
|
How many bonds between cytosine and guanine?
|
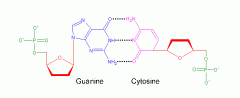
3 Hydrogen bonds!
|
|
|
How many bonds between thymine and adenine?
|
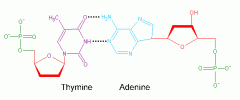
2 Hydrogen bonds!
|
|
|
Where do two nucleotides bond to form a nucleic acid?
|
The phosphate group attached at the 5'-Carbon of one sugar attaches to the 3'-Carbon of the next sugar.
|
|
|
What is the structure of DNA?
|
Complimentary and antiparallel, so one chain runs 5'-3' from top to bottom whereas the other chain runs 5'-3' rom bottom to top.
|
|
|
What is the structure of RNA?
|
Single stranded, runs 5'-3'
|
|
|
What is central dogma?
|
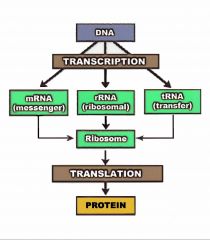
The division of genetic information flow into three stages:
Replication, where DNA is duplicated Transcription, where information from DNA is transferred to RNA (mRNA, tRNA, or rRNA) Translation, where information in RNA is used to build polypeptides |
|
|
What does replication?
|
Enzymes called DNA polymerases
|
|
|
What does transcription?
|
Enzymes called RNA polymerases
|
|
|
What does translation?
|
Ribosomes
|
|
|
What is genetic code?
|
Another aspect of central dogma where genetic information is stored in DNA sequences as 3 nucleotides being decoded at a time. Each 3 DNA nucleotides in a gene code for one of the 20 amino acids.
|
|
|
What is regulation?
|
Another aspect of central dogma that turns genes on to be transcribed only when needed.
|
|
|
What are codons?
|
Three letter sequence that codes for an amino acid.
|
|
|
What are inducible genes?
|
Genes that are not on all of the time. Can be turned on by inducers.
|
|
|
What are constitutive genes?
|
Genes that are always turned on!
|
|
|
What is DNA hybridization in the laboratory?
|
Using the complimentary ability of two strands of DNA to base pair in the same solution if cooled slowly. The basis of many modern diagnostic tests.
|
|
|
What is supercoiling?
|
Allows the DNA to be packed into the cell.
|
|
|
What is negative supercoiling?
|
When the DNA is twisted in the opposite direction of the helix.
Most common in nature. |
|
|
What inserts negative supercoils in DNA?
|
Gyrase, a topoisomerase, or Topoisomerase II.
|
|
|
What is positive supercoiling?
|
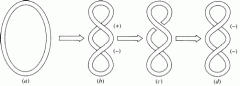
When DNA is twisted in the same direction as the helical turns (right handed).
Examples have been found in extreme thermophiles. |
|
|
What inserts positive supercoils?
|
Reverse gyrase.
|
|
|
What removes supercoiling?
|
Topoisomerase I, which is an enzyme that cleaves the phosphodiester bond in a single strand in the DNA and causes the tension to relax.
|
|
|
What inhibits DNA gyrase?
|
Nalidixic acid, novobiocin, and ciprofloxacin (antibiotics)
|
|
|
What are stem-loop structures?
|
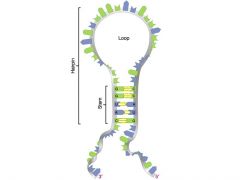
They are formed by inverted repeats which can be inverted and read the same way and cause DNA to assume a structure with unpaired loops between the two repeats.
|
|
|
What are the requirements for DNA replication?
|
1. Template: must have something to copy.
2. DNA Helicase: the is a DNA unwinding protein. 3. Single-stranded binding protein: proteins that bind to the unwound (ss) DNA and prevent it from re-base pairing. 4. Primer: must have a starting place. A short strand of RNA. 5. Primase: a specific type of RNA polymerase that copies the first 11 nucleotides by making an RNA chain 11 ribonucleotides long. 6. DNA polymerase III: enzyme that catalyzes the addition of the nucleotides to the RNA primer and then to the growing DNA strand. 7. Deoxynucleotide triphosphates: dATP, dTTP, dCTP, dGTP. Precursors for the growing chain. 8. DNA polymerase I: When replication is well under way, this enzyme chews up the RNA primer and replaces it with DNA. 9. DNA Ligase: Attaches the ends of two DNA strands. |
|
|
What is DNA Helicase?
|
The DNA unwinding protein
|
|
|
What is primase?
|
A specific type of RNA polymerase that copies the first 11 nucleotides by making an RNA chain 11 ribonucleotides long.
|
|
|
What is DNA polymerase III
|
AN enzyme that catalyzes the addition of the nucleotides to the RNA primer and then to the growing DNA strand.
|
|
|
What is DNA polymerase I?
|
When replication is well under way, this enzyme chews up the RNA primer and replaces it with DNA.
|
|
|
What is DNA ligase?
|
Attaches the ends of two DNA strands.
|
|
|
What is the first step of DNA replication?
|
INITIATION, which begins at the origin of replication.
A. Origin binding protein (DnaA) binds to the origin and then helicase binds and separates the double strands. B. Single-stranded binding proteins bing and prevent strands from repairing. C. Primase makes a short piece of RNA on the leading stand and DNA polymerase III starts synthesizing DNA. D. Primase makes an RNA primer in 5' to 3' direction of the lagging strand. E. Replication begins on the two strands forming the replication fork. |
|
|
What is the leading strand?
|
The strand fowing from the 5' to the 3'. DNA synthesis occurs here continuously because there is always a free 3' -OH at the replication fork to which a new nucleotide can attach.
|
|
|
What is the lagging strand?
|
The opposite strand of DNA where synthesis occurs discontinuously because there is no 3' -OH group.
|
|
|
How does the lagging strand replicate?
|
Through Okazaki fragments.
These are small fragments of the lagging strand that have been paired with many RNA primers by primase to provide free 3' -OH groups. These fragments are later joined together to give a continuous strand of DNA. |
|
|
What is the second step of DNA replication?
|
ELONGATION
A. DNA polymerase III adds DNA to the primer in the 5' to 3' direction. B. DNA polymerase I degrades the RNA primer and replaces it with DNA. |
|
|
What is the third step of DNA replication?
|
TERMINATION
A. The polymerases converge on a region of the DNA containing sequences known as ter sites where they meet and complete the replication process. B. The single DNA strands are joined by DNA ligase. |
|
|
How does DNA proofread to keep the error rate very low (1 in 10^6 to 1 in 10^9)?
|
1. Base pairing: DNA polymerase III reads the template nucleotides accurately and usually inserts the correct complementary nucleotide.
2. DNA polymerase III is able to detect incorrect nucleotides that are not complimentary and remove them by chewing back the new strand from the 3' end (3' exonuclease activity). <--- PROOFREADING. All prokaryotes, eukaryotes, and viruses have this function! |
|
|
What is mismatch repair?
|
By endonucleolytic proteins that are able to detect bases that are mispaired. They excise the mismatched base and replace it with another.
|
|
|
What are the three types of RNA?
|
1. Messenger RNA (mRNA)
2. Transfer RNA, carries the amino acids-adapter molecule (tRNA) 3. Ribosomal RNA, what ribosomes are made of (rRNA) |
|
|
What catalyzes transcription?
|
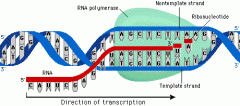
RNA polymerase, a complex protein composed of 5 peptide subunits.
|
|
|
What is the first step of transcription?
|
INITIATION
Where the sigma factor finds the promoter (transciption on/off switch) and the core enzyme binds to the DNA and begins transcribing (making an RNA copy) |
|
|
What is the second step of transcription?
|
ELONGATION
Where the sigma factor dissociates, core enzyme moves down the DNA separating the strands to allow base pairing between the ribonucleoside triphosphates. Only one strand is read. |
|
|
What is the third step of transcription?
|
TERMINATION
The polymerase reaches a sequence in the mRNA called the terminator and falls off the DNA. |
|
|
What are the two types of termination in transcription?
|
1. Rho Dependent: It is a protein that travels behind the RNA polymerase on the NEWLY MADE RNA and when the polymerase reaches a certain Rho utilization termination sequence, it slows down, causing the Rho protein to catch up and knock it off the DNA.
2. Rho independent (intrinsic): inverted repeats form large hairpin structures in the newly made RNA which are followed by a string of uracils,causing it to pair weakly with the template and dissociate. |
|
|
What are operons?
|
Prokaryotic transcriptional unites whose expression is controlled by a single regulator sequence (promoter, operator...) and allows their expression to be coordinate into a single polycistonic mRNA.
Not found in eukaryotes. |
|
|
What are the components of Translation?
|
1. aa-tRNA: carries the amino acids
2. mRNA: carries the genetic instructions from the DNA 3. Ribosomes: catalyze assembly of the amino acids into peptides 4. Factors: proteins that are not part of the ribosome but are required for proper protein synthesis |
|
|
What is the start codon?
|
AUG
It encodes a chemically modified methionine when found at the beginning of a coding region. |
|
|
What are stop codons?
|
UAA
UAG UGA Signal the termination of translation. |
|
|
What is the peptide bond in between two amino acids?
|
A link between the -OH group of one amino acid and the H-NH group of another. Formed by removal of water.
|
|
|
tRNA synthetase?
|
Links tRNA and its specific amino acid together.
Ensures that a particular tRNA receives the correct amino acid and must thus recognize both the tRNA and its cognate amino acid. |
|
|
What is the wobble position?
|
Where base pairing is more flexible for the third base of the codon that for the first two. Leads to degeneracy of genetic code.
|
|
|
How can the reading frame be affected?
|
If the mRNA is read in the right position (0), the proper protein is made.
The other two possible reading frames are a +1 or -1 shift. |
|
|
What are the three binding sites for ribosomes on tRNA?
|
APE
A-site: Aminoacyl site where incoming aa-tRNA enters the ribosomes. P-site: Peptidyl site where the tRNA containing the growing peptide chain is located. E-site: Exite site where the P-site tRNA goes after it has given up its peptide to the tRNA in the A site. |
|
|
What are polysomes?
|
mRNA/ribosome complexes which have several ribosomes translating at the same time.
|
|
|
What happens when a defective mRNA lacking a stop codon stalls a ribosome?
|
tmRNA comes in which has tRNA-Ala and mRNA and creates a peptide tag.
|
|
|
What is the primary structure of proteins?
|
The sequence of amino acids in a peptide or protein.
|
|
|
What is the secondary structure of a protein?
|
The repeating conformational patterns formed by twist and fold in a polypeptide.
Formed by hydrogen bonds between atoms in the peptide bonds, not ones in R-groups. |
|
|
What are chaperonins?
|
Proteins required for proper folding or for assembly into larger complexes.
|
|
|
What are the two possible destinations for protein export?
|
Protein contains SecA and is secreted into the periplasm.
Protein contains signal recognition particle and is inserted into the membrane. |

