![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
23 Cards in this Set
- Front
- Back
|
Genetic Model for Continuously Distributed Traits
|
- Continuously distributed traits are assumed to be MULTIFACTORIAL; influenced by many different factors. Some of these factors = (a) different genes, some will be (b) non-genetic factors (including chance, climate, and nutrition) called environment.
Phenotypic value (performance of an individual for a particular trait) is made up of a genetic component "G" (genotypic value) and an environmental component "E" (environmental deviation). G & E are assumed to be independent of each other. Phenotype = Genotype + Environment P = G + E |
|
|
Breeding Value & Equation for Genotype
|
Breeding Value (A) = The average effect of the gene. (Breeding value is important to be able to identify animals with the best genes, for additive effects. This is significant since it is known that certain gene interactions exist.)
Genotypic Value = Breeding Value (Additive effect effect of gene, "A") + Deviation due to dominance ("D") + Deviation due to interaction b/w genes at different loci (Epistatic deviation "I") G = A + D + I |
|
|
Genotypic Value Equation; significance for selective breeding.
|
G = A + D + I
G = Genotypic value A = Breeding value, additive effect D = Dominance deviation I = Epistatic deviation The most important component for selective breeding is the BREEDING VALUE / ADDITIVE GENETIC COMPONENT. (However, dominance and epistatic components are considered in crossbreeding schemes.) |
|
|
Heritability (in the Broad Sense); H^2
|
Broad-Sense heritability (H^2) = The proportion of variance that is due to genotypic values.
Heritability (broad sense) = VG / Vp VG = Genotypic variance Vp = Variation in phenotype --> (More specifically...) Broad-sense heritability reflects all the genetic contributions to a population's phenotypic variance including additive, dominant, and epistatic (multi-genic interactions), as well as maternal and paternal effects, where individuals are directly affected by their parents' phenotype (such as with milk production in mammals). |
|
|
Heritability (in narrow sense); h^2
|
Narrow-Sense Heritability (h^2) = The proportion of the variance that is attributable to variance in breeding values. It predicts the response to selection, and is the term most valuable to animal breeders.
Heritability (narrow-sense) = VA / Vp VA = Variation in breeding values Vp = Variation in phenotype --> (More specifically...) Narrow-sense heritability captures only a portion of the variance due to additive (allelic) genetic effects. Additive variance is important for selection. If a selective pressure such as improving livestock is exerted, the RESPONSE of the trait is directly related to narrow-sense heritability. |
|
|
Trait Heritability vs. Breeding Value
|
Heritability of a trait and the breeding value of an animal can vary b/w populations, depending on...
(1) gene frequencies, (2) type of gene action, and (3) environmental effects. |
|
|
Response to Selection Equation
|
Response to selection (R) = the realized average difference between the parent generation/current population and the next generation
R = h^2 S h^2 = herritability (narrow-sense) S = the Selection Differential; the strength of selection, the average difference in mean trait between the population and the selected parents of the next generation EXAMPLE: Traits with less heritability, respond slower to selection (with an exception for parasitic disease). The mean of the trait will increase in the next generation as a function of how much the mean of the selected parents differs from the mean of the population from which the selected parents were chosen. The observed response to selection leads to an estimate of the narrow-sense heritability (called realized heritability). This is the principle underlying artificial selection or breeding. [edit] |
|
|
Estimated Breeding Value (EBV)
|
Estimated Breeding Value (EBV) = is a statistical numerical prediction of the genetic value relative to the mean of an individual (male or female) available for breeding. EBVs are used to rank breeding stock for selection, based upon an individual's genetic value for specific traits. An EBV therefore is a calculated estimate of heritability for each trait being considered, relative to the EBVs of other individuals in the breeding population.
Involves a Regression; statistical technique looking at the relationship b/w two variables --> Used to express the phenotype value relative to the mean. |
|
|
Estimated Breeding Value: REGRESSION
|
A regression (which looks at r/ship b/w 2 variables) is used to compare phenotype (x-axis) with breeding value (y-axis). It expresses phenotype relative to the mean.
EXAMPLE: A better than average trait = above the mean. Most traits, above the mean is an improvement (unless it's something like somatic cell count). --> There tends to be less variation in fitness traits than in production traits. |
|
|
Calculating Estimated Breeding Values; Single Measurement
|
If there is ONLY a single record on one animal and NO information on any of his relatives, then the ESTIMATED BREEDING VALUE is the heritability multiplied by the difference between the individual observation and the population mean
EBV = heritability x (difference individual & population mean) EXAMPLE: Lamb = post-weaning growth rate of 500 grams per week, compared to flock mean of 200 grams per week & heritability is 0.33. --> EBV = 0.33 x (500 - 200) = +100 grams/week |
|
|
Estimated Breeding Values: Accuracy & Reliability
|
With a single record, the accuracy and reliability = relatively low. Additional info is usually used; (a) repeated records, (b) info on relatives (especially sibs, offspring, or parents). EBV's usually are calculated by specialized computer programs that take in all available info into account. The more info available = more reliable and accurate the results are.
ACCURACY (of EBV) = correlation b/w the true and the predicted breeding value. --> For a single measurement, accuracy = the square root of the heritability. RELIABILITY (of EBV) = the square of the accuracy. |
|
|
Estimated Breeding Values; Regression Slope
|
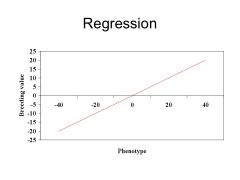
Regression Slope; a line depicting the relationship b/w phenotype (x) and breeding value (y), and can be used to predict breeding value.
THEORY OF REGRESSION; the slope of the line is measured by the covariance b/w breeding value & phenotype, divided by variance of phenotype. --> Regression Slope = Covariance (a, p) / Variance p From theoretical calculations, can estimate heritability from the predicted breeding value... Covariance (a,p) = Variance a B/c assume no r/ship b/w genotype and environment, p breaks down to a + d + i + e, and breeding value is independent of d, i, e (dominance, epistasis, environment) So... Regression Slope = variance (a) / variance (p) = h^2 |
|
|
Estimated Breeding Values; Regression Coefficient Factors
|
Regression Coefficient = Slope
Depends on: (1) Heritability (2) Number of measurements (3) Repeatability of measurements (4) Number of relatives (5) Genetic relationships --> Some traits provide more information than others; the ones that aren't repeatable = the ones that provide more genetic info (tests that are repeatable = usually are less useful). --> An individual's genes get halved to successive generation. |
|
|
Repeatability
|
Repeatability describes the tendency for animals to maintain their production ranking over time. It is therefore a measure of the accuracy with which early records of an animal's performance in a particular trait can predict its lifetime performance.
It indicates the usefulness of repeated records; indicating whether there is value in taking repeated measurements as the basis of selection decisions for the current flock. Repeatability is used to assess which individuals to cull and which to keep in terms of lifetime performance. However, repeatability is not a genetic characteristic, so offers little useful information about selecting individuals as parents. |
|
|
Repeatability; EQUATION
|
The variance b/w individuals (Vb) divided by total variance--the sum of variance b/w individuals and the variance w/in individuals (Vb + Vw)
Repeatability = Vb / (Vb + Vw) Values range b/w 0 and 1. Two or more observations are needed per individual. |
|
|
High and Low Repeatability Values
|
LOW: Low repeatability value = means the same individual can vary widely, not a particularly reliable test.
--> Repeatability decreases as repeated records become less similar; as the amount of variation w/in individuals increases. HIGH: High repeatability = little value in taking replicate measurements. --> When most of the variation is among individuals. |
|
|
REPEATABILITY: Equation (w/ Environmental Variance)
|
Repeatability can be thought of as...the sum of genotypic variance and permanent environmental variance, divided by phenotypic variance.
( Vg + Vep ) / Vp Meaning, the similarity between observations depends on the genotypic component plus the permanent environmental component. The repeatability should be equal to or greater than the heritability. --> Since repeatability can be quickly estimated (even w/out pedigree records), it provides a convenient way to estimate the upper limit of the heritability. (If have a low repeatability, it's bound to have an even lower heritability. --> Remember p = g + pe + te (phenotype = genotype + permanent environment effect + temporary environment effect). The temporary environment effect = likely whats going to change b/w measurements. (Whereas permanent environment & genotype won't change b/w measurements.) |
|
|
Genetic correlations
|
If two traits = genetically correlated, it means their breeding values are correlated;
(a) the value for one trait can be predicted from the other. (b) Selection for one trait would also produce a change in the genetically correlated trait. -->Selection can increase one trait while leaving the other unchanged, or achieve more rapid progress in both. --> W/ neg. genetic correlation (milk prod'n & protein %) it is possible to increase both traits, but progress is slower. (c) Difficult to estimate precisely. --> Values vary b/w -1 (events negatively correlated) to +1 (positively correlated). (d) Important for selection index, indicator traits (correlated w/ economically important traits) |
|
|
Indicator Traits
|
Those traits that are relatively unimportant in themselves, but are genetically correlated w/ economically important traits.
Can be used to select for traits that are difficult to measure or have low heritability. --> Somatic cell counts are an indicator of mastitis incidence and they are being measured in future selection indices. Selection will then either reduce the incidence of mastitis, or prevent it rising. |
|
|
Genetic Correlations: PERFECT CORRELATION
|
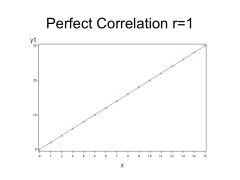
PERFECT Correlation, is r = 1
--> Perfect correlations are seldom seen in Biology. |
|
|
Genetic Correlations: HIGH CORRELATION
|
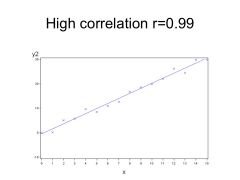
HIGH Correlation would be close to 1, r= 0.99
|
|
|
Genetic Correlations: MODERATE CORRELATION
|
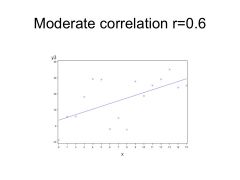
Quite a bit of spread amongst data. MODERATE CORRELATION might be around r = 0.6
--> Correlation at r = 0.3 ; indistinguishable w/ something w/ NO correlation (w/out statistical evidence) |
|
|
Genetic Correlation; EXAMPLES
|
(1) Milk Yield & Milk Solids; (Individual bulls w/ daughters producing MORE milk, will have more protein & fat b/c of increased milk yields.)
(2) Milk Yield & Mastitis; (Higher milk prod'n increases likelihood of mastitis) (3) Somatic Cell Counts & Mastitis; (SCC can be used as indirect indicator of mastitis. However, if select based on SCC, you may make decisions to decrease a cows ability to respond to infection...) --> Genetic correlations affect how we breed, & important breeding decisions. |

