![]()
![]()
![]()
Use LEFT and RIGHT arrow keys to navigate between flashcards;
Use UP and DOWN arrow keys to flip the card;
H to show hint;
A reads text to speech;
36 Cards in this Set
- Front
- Back
|
only the ______ anomer sugar is observed in DNA
|
β
|
|
|
DNA synthesis always proceeds in the _____ to _____ direction
(parent strand is read in ___ to ___ direction) |
5' to 3'
parent read in 3' to 5' direction |
|
|
what is the correlation between # of DNA base pairs and # of chromosomes?
|
↑ bp ≡ ↑ chromosomes
|
|
|
what is the correlation between # of genes and # of chromosomes?
|
no relationship
|
|
|
What occurs in reverse transcription?
|
Reverse transcriptase performs a three stage conversion of RNA into DNA (RNA virus)
1. synthesis of DNA complimentary to RNA (cDNA) 2. digestion of RNA 3. synthesis of 2nd strand of DNA |
|
|
exons
introns |
exons- the expressed sequences of a gene
introns- the intervening (non-expressed) sequences |
|
|
salvage pathway
|
5-phosphoribose-1-PP (PRPP) + base −> nucleotide
|
|
|
what drug is a potent inhibitor of Thymidylate synthase?
|
prodrug Fluorouracil -> Fluorodeoxyuridylate
|
|
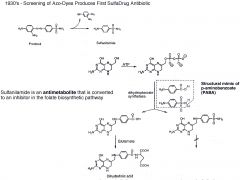
Why was Prontosil screening successful?
|
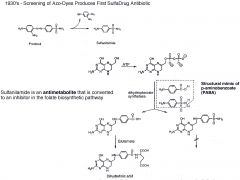
It was screened on mice, not on a plate. It would have failed in plate b/c it needs to be reduced to Sulfanilamide in the liver.
Sulfanilamide mimics the substrate PABA in the folate pathway. |
|
|
What is the function of Mg in DNA synthesis?
How many are needed and why? |
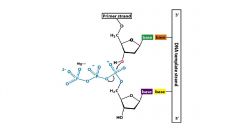
Mg neutralizes Pi, making a better leaving group and allowing nucleophiles to attack the phosphate of interest
Need 2 Mg++: 1. coordinates 3' -OH; increases acidity of proton and allows formation of nucleophillic alkoxide 2. coordinates dNTP phosphates neutralizing charge and providing scaffold for replacement reaction |
|
|
Which Watson-Crick base pairing is stronger and why?
|
G-C bonds are stronger because they have 3 hydrogen bonds vs. A-T have 2
|
|
|
exonuclease
|
cuts a nucleotide off the end of DNA strand
|
|
|
endonuclease
|
cuts into the middle of a DNA strand to remove a nucleotide
|
|
|
Okazaki fragments
|
newly synthesized DNA segments on the lagging strand
- small fragments are part of the stepwise replication of the lagging strand. Followed by pol I removing the primer then ligase joining the strands together |
|
|
What is the mechanism of Topoisomerase I?
Does it use ATP? |
Topoisomerase I relaxes the DNA by using an active site Tyrosine to cause a nick in DNA, then holds close to be put back together after relaxing
- does not need ATP |
|
|
What is the mechanism of Topoisomerase II?
Does it use ATP? |
Topoisomerase II breaks both strands of DNA so another strand can pass through, then the strands are put back together
- needs ATP - would like to have this for untangling a fine metal chain; could just pop a strand right out of a knot |
|
|
What is the replication rate of DNA polymerase III?
|
500 nucleotides per second ( x2 strands → 1000/sec)
|
|
|
Helicase:
What does it do? How does it do it? Does it use ATP? |
Helicase splits and opens a DNA strand for replication. AND it recruits the other enzymes needed for replication.
It spins and uses a ratchet mechanism (grab, pull, release) to slide along one of the strands. Conformational change of the 'ratchet' occurs with ATP binding and hydrolysis. Yes, it uses ATP |
|
|
DnaB
|
helicase
- the 'ratchet' that separates strands of DNA for replication |
|
|
What is the purpose of the RNA primer in DNA synthesis?
Is it accurate? Why/why not? |
DNA polymerase requires the primer to start
It is not very accurate. It does not need to be because it is temporary. |
|
|
What is the structure and mechanism of the 'sliding clamp' on DNA polymerase III
|
1. Opened clamp attaches to lagging ssDNA
2. Lots of α-helices on inside project pos. charge (i.e. Arg) to lightly bind enough to stick but allow sliding |
|
|
What is the inhibitor of de novo Purine synthesis?
|
allosteric regulation of Aspartate Transcarbamoylase by CTP
|
|
|
What are the inhibitors of Pyrimidane Synthesis?
|
1. IMP, AMP, & GMP inhibit formation of Phosphoribosylamine from Ribose 5-phosphate
2. AMP inhibits formation of Adenylosuccinate (AMP precursor) from IMP 3. GMP inhibits formation of Xanthylate (GMP precursor) from IMP |
|
|
DNA polymerase I
|
erases primer and fills in gaps on lagging strand
|
|
|
DNA polymerase II
|
DNA repair
|
|
|
primase
|
synthesizes the RNA primer at the beginning of DNA replication
|
|
|
DNA Polymerase III
|
the workhorse of DNA replication
- quickly synthesizes the leading strand in one sequence - synthesizes Okazaki fragments of the lagging strand - many DNA pol III's include a proofreading mechanism which decreases errors by 100 fold |
|
|
Topoisomerase IV
|
separates genomic DNA molecules after replication
|
|
|
What drug is an inhibitor of Topoisomerase II
|
Cipro
|
|
|
Transition
|
purine to purine or pyrimidine to pyrimidine mutation
|
|
|
Transversion
|
purine to pyrimidine or pyrimidine to purine mutation
|
|
|
What does the 'finger domain' of DNA pol III do?
|
binding of correct nuceotide triggers a conformation change that aligns the substrates and increases probability of reaction
incorporation of incorrect base leads to conformational change and base removal by exonuclease activity (hydrolysis of phosphodiester bond) |
|
|
In Watson-Crick base pairing, H-bonding is important for ____________, but less important for ____________
|
H-bonding is important for duplex stability
polymerase just looks at shape, not at h-bonds |
|
|
How do bacterial and eukaryotic replication differ regarding concurrent replications?
|
bacteria- one replication bubble
eukaryotes- many replication bubbles |
|
|
How does DNA replication know where to start?
|
origins have a unique sequence structure "oriC" (a tandem array of 13-mer sequence) where DnaA binds then recruits the other enzymes
|
|
|
How are the ends of DNA protected from degredation?
|
telomerase adds short, repeat telomere sequences to the ends of chromosomes
|

